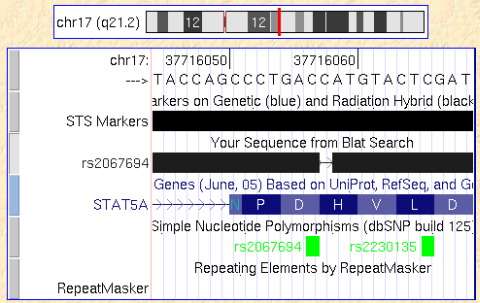
Here I describe a bug in the UCSC Genome Browser display that I found this morning. When locating a SNP on the human genome (where I have only surrounding sequence) I usually do a quick BLAT search of my sequence with the SNP base being deleted. This will show up as 1 bp gap that can be easily matched to exon-intron structure or reading frame. Occasionally, however, there is a +1 bp offset of my artifical gapwhich would let me draw wrong conclusions. My BLAT search below has the name “rs2067694” and shoudl match the SNP indicated in the bottom line.
If you want to use my settings, cut & paste “http://www.wjst.de/wjst_snp.ini” into this linked page.