So far in epidemiology case – control studies are defined by an approach where
… the past histories of patients (the cases) suffering from the condition of interest are compared to the past histories of persons (the controls) who do not have the condition of interest, but who otherwise resemble the cases in such particulars as age and sex ….
I usually explain controls as non-cases in the same overall environment – having same age + sex + geographical area is always a good choice for controls. If there is a known strong risk factor (like smoking for lung cancer) I would also match controls for this risk factor.
This concept now gets heavily modified when reading the recent Nature GWA papers. Here the authors use two control groups (as they feared a possible ascertainment bias) but judge this later as a minor problem as including principal component factors did not lead to an overall change of the results.
Nevertheless there could be regional stratification of certain alleles (as evident from table 1) that may have affected single SNPs – and let me wonder why the main SNPs associations have not been explicitly tested for stratification.
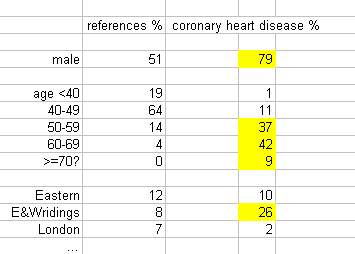
Look at supplemental table 1:
26% of all coronary heart disease cases come from E & W Ridings but only 8% of the controls. Any allele more prevalent in E & W Ridings will pop up more frequently in the cases leading to a spurious association. Unfortunately, there are even more uncomfortable effects:
One consequence of using a shared control group (for which detailed phenotyping for all traits of interest is not available) relates to the potential for misclassification bias: a proportion of the controls is likely to have the disease of interest (and therefore might meet the criteria for inclusion as a case) and some others will develop it in the future. However, the effect this has on power is modest unless the extent of misclassification bias is substantial; for example, if 5% of controls would meet the definition of cases at the same age, the loss of power is approximately the same as that due to a reduction of the sample size by 10%.
This can also found in the table above. Coronary heart disease has a prevalence of about ~5% and most of the controls are too young for manifestation of the disease. We are therefore further underestimating effects – I believe even to a larger extent than the authors make us believe. This will not cure any hidden stratification effect as it will operate on different SNPs.
A further problem relates to the differences in sex distribution between cases and controls. By running an analysis of coronary heart disease we are comparing in 29% of the sample men with women. As this is not a genetic study alone (it needs also environmental factors for expression of the reported diseases), this adds further uncertainty. Case will have bad genes + damaging environment, controls are either missing genes or environment (or both), probably leading to a further underestimation of effects.
Why not calling this a case-reference study rather than a case-control study?
Unfortunately, there is not so much that could be done now about false negative results – they have simply been missed. The only thing we could do is to look at replication of previous results of which 14 SNPs are being listed in table 2. We may immediately subtract the first SNP – simply not associated (1.7×10-1) as well as the 7th SNP – not tested. It is a matter of choice if we want to include the 2 HLA SNPs here as they are likely introduced here by stratification. Don´t ask the question if a hit at or close to a gene will also count as replication when r2 is zero, as there will not so much remain in the basket.
Will these studies ever been repeated with adequate controls?