I have now used the PLINK software package also for analysing family data. Again it is an excellent program that allows many new views on old data. I could run >20 traits all together on ~1000 SNPs in just a few seconds. There are only a few tweaks that I have already emailed to the author like
–hap-tdt option requires the flag –old-EM
–all-pheno option works for QTLs but not for categorial phenotypes
–lookup option needs to have its own output file
–epistasis option should work in families
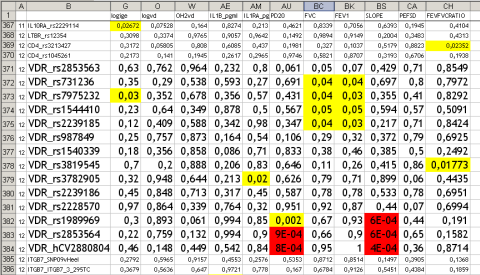
The “downside” – there are so many new results from the German asthma family study, mostly additions to already published papers that will be difficult to re-publish. For example VDR variants influence also lung function and bronchial hyperreactivity which has not been tested before.
BTW – the title of this post goes back to an article in JSTOR