I am struggling now for 4 weeks with some unusual behaviour in PLINK that gives me different results with a trait of the alternate phenotype files either by calling that trait directly
plink –file mydata –tdt –pheno pheno2.txt –mpheno 1
or from a loop over all traits
plink –file mydata –tdt –pheno pheno2.txt –all-pheno
It seems that I am working with different numbers at both occasions – click to enlarge the log
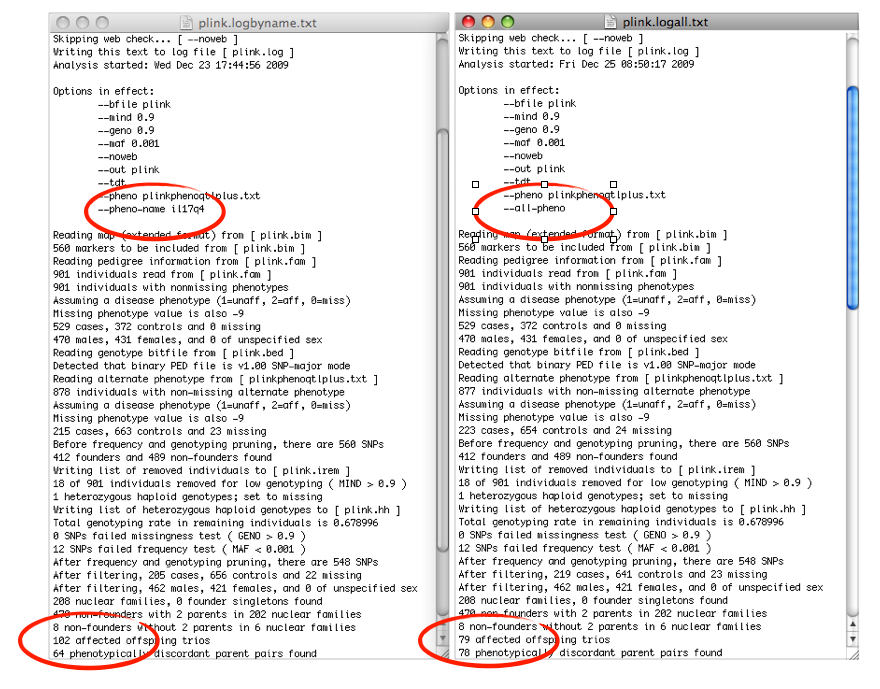
Case numbers in the –all-pheno output above are probably misleading (as case definition is changing during every loop run), while the main problem are different transmission counts of the TDT (there is no problem with the –assoc option).
Playing around with the data it turned out that changing a few observations in the FIRST phenotype alters also the results of a CONSECUTIVE trait.
I have no response from the author so far while peeking into the source code did not help so much. A workaround would be to avoid the –all-pheno option and use a bash loop
for i in eotaxin gmcsf ifng il4 lasttrait
do
/Applications/Genetics/plink-1.07-mac-intel/plink –bfile plink –mind 0.9 –geno 0.9 –maf 0.001 –noweb –out $i –tdt –pheno plinkphenoqtlplus.txt –pheno-name $i
done