The Rosetta stone (I took the picture above earlier this year in the British museum) has become the key to decipher Hieroglyphic as it contained the same text also in Demotic Egyptian and Greek. Discovered by a French in 1799, brought to England in 1802 it become eventually translated in 1822 by Jean-François Champollion.
Continue reading The Rosetta stone and the genetic code
Category Archives: Genetics
Ethnographic studies at Oktoberfest
Having many years of experience with ethnographic studies at Oktoberfest München, I am fascinated by a new Cell paper that shows distinct behavioral responses to ethanol. This is something that I alread assumed (although I did not known about this particular RhoGAP18B isoform only about ADH deficiency). Will the knowledge of more and more mutations in the lifestyle area raise ethical problems? Yea, yea.
Once again genetic testing
I have argued earlier that the free decision of an individual to allow genetic testing, will also reveal data on genetic relatives that have never consented to that procedure.
A new review by Bruce Weir now confirms that “it is reasonably straightforward to find the probability of the genotypes of individuals when their relationship is known…” My current work lets me also assume that with 500,000 SNP data at hand, much individual characteristics of the donor can be reconstructed – there are no anonymous DNAs datasets as some people still believe.
I even fear that genetic testing will increase for example in “homeless” (in vitro fertilized) individuals as these people will want to prevent sibling marriage – see for example the a-China DNA project. Other people may be curious about their genealogy, others about drug side effect prediction, lifestyle, assurance questions…
With every new dataset, available datasets will gradually decrease their anonymity level. I fear that anonymity is not so much a dichotomous property, it is much more a likelihood ratio to stay unknown under the probability to be known. Yea, yea.
Addendum
Time online of Dec 17, 2006 reports that the British police is holding the DNA records of more than 1m innocent people — eight times more than ministers have previously admitted. I wonder if this will affect participation rate of the UK Biobank that targets health of lifestyle, environment and genes in 500,000 people.
Sleep well
Just came to my attention that there is research of sleep related genes, the usual stuff of protein kinases, dopaminergic receptor, and serotonine transporter. Also this research community seem to have the common difficulty of the complex disease gene mappers – to understand a phenomenon (not a trait) as systemic function, an intrinsic property of a multicellular and multiwired brain, yea, yea.
Addendum
More strange phenotypes orgasm frequency, pain, human memory performance.
Addendum
Read also the what-we-could-have-learned-from-linkage-studies.
Heritable mutation or not?
moblog – Tsun Leung Chan is now reporting a heritable germline epimutation of MSH2 in a family with hereditary colorectal cancer another case of “paranormal” inheritance. They find a mosaic germline methylation pattern (which might even be a symptom of another mutation that affects the demethylation-de novo methylation pattern of MSH2 during embryogenesis?). If my hypothesis is true these families should even show more genes with different methylation patterns, yea, yea.
Addendum 28/5/08
Another attempt to answer this question comes by a study of the MLH1 promotor
Pro: MLH1 promoter methylation was found in a patient and his mother giving evidence for a familial predisposition for an epimutation in MLH1. Contra: a de novo set-up of methylation in one patient, a mosaic or incomplete methylation pattern in six patients, and no evidence for inheritance of MLH1 promoter methylation in the remaining families.
What could have been learned from linkage studies
What makes the difference between genetic linkage and association studies? Simply speaking, for linkage you need to inherit a particular marker allele from your parents where it does not matter if a child in another family inherits another allele (pending it shares it with its affected sibling). With association studies this matters.
As we found with the much relaxed linkage strategy so many minor diverse loci, I assume a rather heterogeneous origin of complex diseases. There is no doubt about the importance of genes, but about the sharing of the same genetic abnormality. An (anonymous) position paper on basic Asthma Research Strategy II in Clin Exp All 2006; 36: 1326 says
The average size of effect on asthma and related traits from common SNPs is small. For instance, seven common SNPs in the IL13 gene jointly accounted for only 0,5% of the variance of total IgE … With a heritability of circa 60% for total IgE this implies that hundreds of genes, each with small effects, may be involved in IgE regulation.
Families presenting with a complex system disease will all have unique patterns how they arrive at the same clinical endpoint. Alpha-delta-gamma asthma, theta-kappa-jota schizophrenia or $%&# diabetes – are they combining lets say 3000 variations in 300 genes of 30 metabolic-signalling pathways to 1 disease of variable onset, severity and prognosis? Yea, yea.
Free for all
A report describing some first experience with GWAs (that means genomewide association studies by single nucleotide polymorphism) is listing several websites that will offer soon public data
- Hapmap [available]
- CEPH [available]
- Affymetrix 500 K Hapmap [available]
- a Parkinson study [available]
- GAIN [forthcoming]
- Wellcome Trust [forthcoming]
- NIH [forthcoming]
- Affymetrix reference set Affymetrix / IKMB Kiel / Erasmus Medisch Centrum / Karolinska Institutet / Centre National de Génotypage [forthcoming]
- CGEMS
There are many opportunities to do research without ever writing a grant application, yea, yea.
Addendum
A German paper gives some updates – you may also check www.p3gconsortium.org and www.wtccc.org.
A zebra takes its stripes wherever it goes
This week ends with another African proverb and some thoughts on transcription factor binding sites.
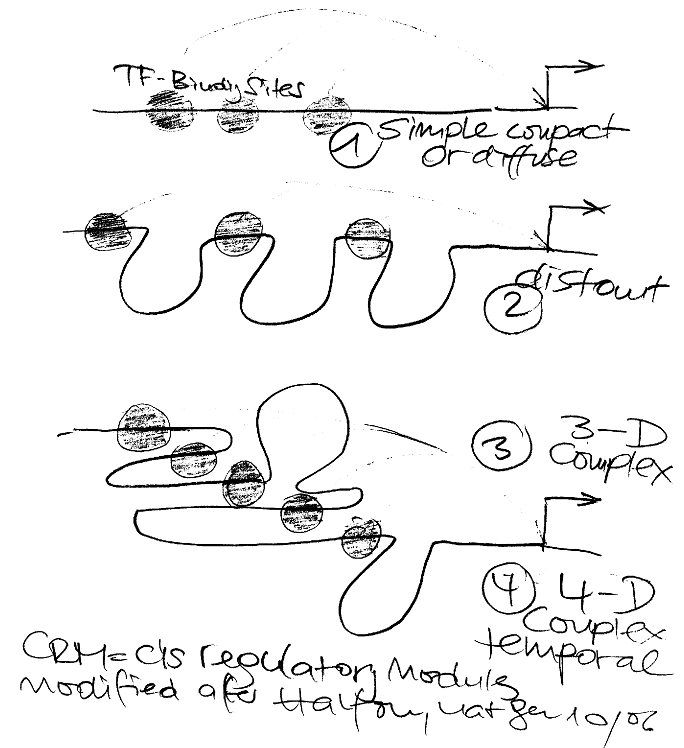
Commenting on the work of Janssens that “there is no inherent reason that CRMs [cis regulatory modules] must lie on a contiguous segment of DNA or that they must contain clusters of sites”, Halfon still expresses the hope that “we should be able to identify the regulatory elements for any gene starting with only the DNA sequence”.
Although an old dream, I am now rather sceptical if this prediction will ever work. Recognize the differences of my drawing compared to original figure: I am introducing even another level of temporal relationships. There might be even a close relationship to yesterdays post on “retaliation“, yea, yea.
Do not run after a cart that will not take you
Starting with another African proverb, here are some thoughts about evolution, design and the difference of chimps and humans. Yes, I am biased, I know.
I have learned that there are mainly three differences between chimps and human – the ability to run, a larger brain size and the language/speech capability. The only trait that can directly observed is the ability to run (check Munich marathon: Neither brain size and language can be directly observed :-) BTW, I renember having seen a family that walk on feet and hands – quadrupedal locomotion is a recessive trait linked to chromosome 17p, the way we all start our lifes).
So genetics is playing a big role in the human < -> ape differentiation. Or did the differentation select the genes?
You will understand my great expectations when now reading one of the first serious papers about the chimp and the human lineage. It is about pseudogenization, the gene loss during separation of species. The authors show 80 non-processed pseudogenes inactivated in the human lineage – while gently negelecting the fact of another 7868 or so pseudogenes in the human pseudogene database.
There is also nothing about my favorite trait bipedalism (only a ridiculous quote of pseudogenization of the sarcomeric myosin gene MYH16 that should relate to hominin masticatory muscles that “may have allowed the brain size expansion”, uhhh. It is also hard to understand how gain of ability should be caused by loss of gene function, yea, yea.
Genetic archaeology
While doing a study of European population stratification, I came across an older but interesting study of old testament priests that compares the Levi tribe (where Moses was a member) with the Cohanim tribe (descendents of Moses’s brother Aaron who served as priests). The investigators traced patrilineal inheritance since the temple period 3,000 -2,000 years until present, and showed current levites unlike the Cohanin having a heterogeneous origin. The coalescence of of Cohanim chromosomes is dated to between Exodus and the destruction of the first temple in 586 BC.
Most current research is dedicated to between species comparisons but unfortunately the wonderful older Y and mtDNA approaches haven´t kept pace with the current SNP technology developement. There would be many intersting studies possible following the timeline of European history, yea, yea.
Father son relationships
This year Noble prize in physiology and medicine honored for the 6th time a famous son of a famous father (the Kornbergs). We have a saying that the apple does not fall so far way. Yea, yea.
Genomics of a single cell
This is a fascinating area – methods for amplifying DNA from single cells for complete genome sequencing. The current approach uses isothermal multiple displacement amplification MDA, followed by 454 sequencing. Biotechnology is getting closer towards its origins, yea, yea.
Addendum
We can now add also single cell protein analysis – microfluidic devices that can dissolve and separate proteins before quantifying them by fluorescence in confocal microscopy. Several groups tried to increase the signal-noise ratio by decreasing capillary dimension (leading to clogging) while the new study widens the excitation laser.
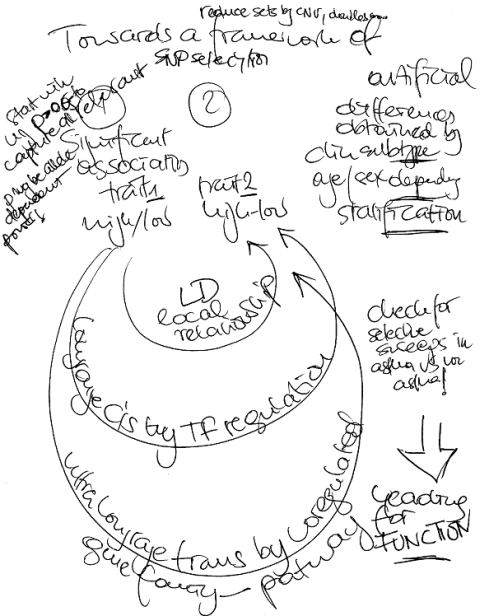
Towards a framework of SNP interaction
The forthcoming SNP whole genome association studies will draw a lot of attention. I already have the problem of many interacting SNPs where I am suggesting to divide significant results in 3 compartments – local (LD effects), longrange (cis transcription factor effects) and pathway effects (gene family, system wide effects). The main problem are false positives that can even not be overcome by techniques like FDR. Yea, yea.
Systems biology
A nature genetics paper shows that a single mutation in Pseudomonas has pleiotropic effects – not limited to the level of proteins in the particular network but changing also their relationships. Sounds like a soccer match where a player has to leave the court – which changes positions of all remaining players. “The adaptive mutation draws proteins into tighter coregulation”, yea, yea.
Bereshit bara Elohim et hashamayim ve’et ha’arets…
The genesis – the common book of Jewish and Christian faith – reports the begining of the world. Interestingly , the creation of living things include also genes in the same instance: “1.11 let the earth spawn grass and leaves that make seeds…” We have been told that reverse genetics may not be used any more. Whats about forward genetics? Yea, yea.