A nice PNAS paper gives in its supplement 4 the MCM6 and mtDNA 16209 genotypes of some prehistoric bones AND of all authors – excellent! The study itself is also interesting as we learn about the true wild-type allele in early Neolithic Europeans – they could not digest milk.
Tag Archives: Genetics + Biology
Conservapedia “gene” entry
Genes and genius
Following many strange gene association studies now a 3rd study of genes and genius appeared: WashU on IQ and CHRM2 believe it or not as part of a study on the genetics of alcoholism…
Dual vitamin D effects on type 1 diabetes and allergy
Early vitamin D supplementation may have dual effects, protecting against type 1 diabetes and inducing allergy. Is that possible? Yes, individual genes variants may determine the outcome.
In addition to HLA the most prominent type 1 diabetes genes are INS – CTLA4 – PTPN22 – CD25 – IF1H1, the most prominent asthma/allergy genes ADAM33 – DPP10 – IL12 – IL4 – GPR154 – FLG. I can´t see any joint master regulator, however, we know that
- CD25 is the IL2R
- and just learned from an excellent new mouse study that IL2 gene variation impairs regulatory T cell function
- while IL2 variants are associated with allergy
- IL2 is suppressed by vitamin D – known since 1985
Could that be a model to explain the dual involvement of vitamin D in both diseases?
Flip-Flop
The AJHG discusses an interesting phenomenon that I noticed too. While replicating a disease marker association the risk allele suddenly reversed which led me believe that there was an error in allele coding. As the authors of this new paper now show these flip-flop association may indeed a true positive association to a non causal variant.
Undo button
–Day 7 of Just Science Week–
Wouldn´t it be nice to have also a CTRL+Z action in the laboratory? For example when you have confused pellet and supernatant during pipetting? Biology at least seems to have some kind of undo action – see a series of nice papers in Cell Research. It´s not about demethylation of the parental genome, it’s about stopping and rebuilding the zygotic transcription program during the first meiotic division (that creates the haploid set of chromosomes that will be passed to the progeny).
At present there are 3 hypothesis around, how transcription is being silenced – simply by the speed up of the cell cycle, by active inhibitory (transcription) factors, or the passive deficiency of critical factors. Sun et al. now show that there is a genome-wide disscociation of chromatin factors leading to a naive state in preparing the new life cycle. Critical transcription factors and regulators remain separated for a prolonged time period and become reassociated only after pronuclear formation. It is still unclear if the second or third hypothesis fits best this process as the absence of even 1 essential transcription factor can inactivate transcription. Deletion of TBP for example will inactivate both PolI and PolII; TBP is found to dissociate among other factors.
Only a couple of structural proteins remain bound persistently (HP1alpha, HP1beta, TOPIIalpha and AcH4, with acetylated histone 4 as a positive control. Of course methyl-binding proteins, topisozymerases, and other heterochromatin binding stuff is required for normal chromatin structure where it would be nice to know which of these remain bound during this reprogramming step. So, this looks more like a reinstalling the OS than a simple undo action.
Thanks and good bye to all guest readers of the science week.
My compliments
My compliments to Nicole, the latest Ph.D. student from our lab who succesfully passed her final exam today in Freising at TU München-Weihenstephan. Here is the semi-official document:
The title of her thesis is High-resolution snp scan of chromosome 6p21 in pooled samples from patients with complex diseases , a topic that has recently attracted new interest.
We apply a high-throughput protocol of chip-based mass spectrometry (matrix-assisted laser desorption/ionization time-of-flight; MALDI-TOF) as a method of screening for differences in single-nucleotide polymorphism (SNP) allele frequencies. Using pooled DNA from individuals with asthma, Crohn’s disease (CD), schizophrenia, type 1 diabetes (T1D), and controls, we selected 534 SNPs from an initial set of 1435 SNPs spanning a 25-Mb region on chromosome 6p21. The standard deviations of measurements of time of flight at different dots, from different PCRs, and from different pools indicate reliable results on each analysis step. In 90% of the disease-control comparisons we found allelic differences of <10%. Of the T1D samples, which served as a positive control, 10 SNPs with significant differences were observed after taking into account multiple testing. Of these 10 SNPs, 5 are located between DQB1 and DRB1, confirming the known association with the DR3 and DR4 haplotypes whereas two additional SNPs also reproduced known associations of T1D with DOB and LTA. In the CD pool also, two earlier described associations were found with SNPs close to DRB1 and MICA. Additional associations were found in the schizophrenia and asthma pools. They should be confirmed in individual samples or can be used to develop further quality criteria for accepting true differences between pools. The determination of SNP allele frequencies in pooled DNA appears to be of value in assigning further genotyping priorities also in large linkage regions.
Genes on the move
–Day 2 of Just Science Week–
Most people think that human genes are static entities inherited from generation to generation. They may be right, there are no jumping genes in humans.
In 2000, when defending my thesis in epidemiology, I even had to answer the question of the faculty: “How can allergy have a genetic cause as most allergy cases date back only 1 or 2 generations?”. I explained the concept of susceptibility genes (that were always there) plus some new environmental risk factor (that came in only recently) and passed the colloquium.
Maybe this concept was not completely wrong. By today, however, I could offer more explanations – human genes are on the move and even within 2 or 3 generations. You may still wonder – are we talking about T cell receptor recombination? Yes, this may be a possibility, but not a really new one. More noteworthy are (1) abolished purifying selection (2) population admixture and (3) increased spike in mutations. These are all are independent paths that may combine freely.
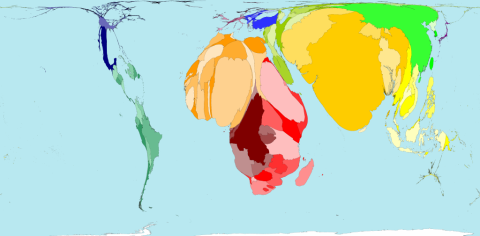
Lets start with “abolished purifying selection”. At the beginning of the last century there were much larger family sizes and a much higher infant mortality. In Europe, mortality under the age of 5 has been about 250/1.000 live born children in 1900. It dropped to 50/1.000 around 1950 and is now about 5/1.000 – the effect of improved sanitary conditions, vaccines and antibiotics. Geneticists would describe it as a reduced selection – giving immediate rise to some variants in the gene pool.
The figure is © Copyright 2006 SASI Group (University of Sheffield) and Mark Newman (University of Michigan). Thanks to John Pritchard from the Worldmapper Team to let me post it here. Territory size shows the proportion of infant deaths during the first year of their life in 2002
Second, think of population admixture. This usually refers to the composition of a population by descendants of a few founders. Except for major migration periods population composition has been kept rather stable over centuries which can be nicely seen in humans living in isolation and having enriched some diseases – examples from Finland, Hutterities, South Tirol, East Adria or Iceand. With current decrease of admixture also the prevalence of diseases frequently seen in these populations will go done. However, some people even expect that other diseases will rise – as unusual allelic variants will meet other unusual allelic variants (which has not been balanced before). This theory is still vague but has interesting aspects that may be followed up.
A third possibility why human gene variants may change within short time comes with the ever increasing age of fathers at reproduction. I came across this only very recently by a paper of Ellegren. With each additional year of the father the number of pre-meiotic cell divisions increases – leading to a permanent (germline) increase of mutations. Most of these will be irrelevant but some may spike in random genes leading eventually to health effects. Crow asks: “Is this a problem? Surely it will be eventually, but probably not immediately”.
Human genes are therefore on the move, yea, yea.
Basic course in sociobiology
Quick link: German FAZ has a series of 18 articles – great.
Gene therapy in jail
sorry, typo. Press telegram reports that the geneticist William French Anderson was sentenced yesterday to 14 years in prison. The reason, however, was not doing premature human experiments but molesting his assistant’s young daughter, yea, yea.
Genes wanted
The NIH and Jackson ask for nominations of their gene targeting approach (see also A mouse for All Reasons and my previous comment on the 3 R)
KOMP is a trans-NIH initiative to generate a public resource of mouse embryonic stem (ES) cells containing a null mutation in every gene in the mouse genome. Both conditional and null knockouts are being generated. The purpose of this form is to gather input from the scientific community on which genes should have the highest priority for being knocked out.
The Cell paper also explains the hard to understand differences in knockouts
- targeted deletion
- targeted conditional
- trapped conditional
although I still have semantic problems to understand the nomenclature. Anyway my whishlist – you can do me a favor by voting for CYP27B1, VDR, CYP24A1, OPN, IL4, IL5, IL10, IL12, IL13, FLG, CCR5 and CCR9.
Addendum
You can also leave some input at the Environmental Genome Project.
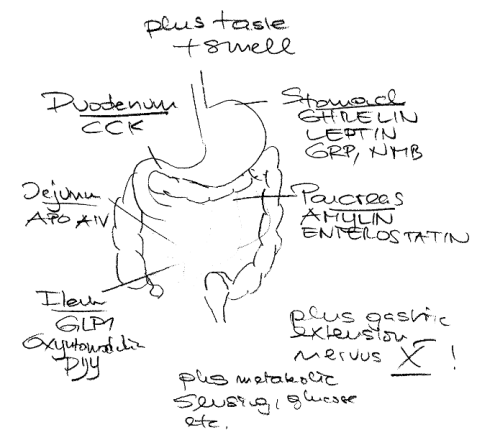
Supersize me II
Some of you may remember the fake food hypothesis that relates the obesity epidemic to the introduction of highly processed industrial food. JCI now has a nice review on the satiation signal and the complex system that may be disturbed.
Microbe content might also be important in this context click | click | click, however, even the adoptive transfer might be a secondary effect – everything in biochemistry follows mass equilibrium constants. The poorer resorption in the lean may lead to a different colonialization and vice versa. Will the adoptive transfer really show lasting effects longer than 2 weeks (in the mouse study)? Isn´t colonialization not influenced by diet (in the human study)?
Yea, yea.
Enter without knocking if you can
This was posted at the door of Max Delbrück (1906-2006) in Pasadena – and quoted from the wonderful biography of E.P. Fischer his last Ph.D. student.
/-/
“Licht und Leben” (or “light and life”) is a wonderful narrative that I really enjoyed, with a lot of informative figures and tables.
I wonder why this biography never appeared in English, why neither institutes in Berlin, Cologne or Constance (where Delbrück teached) are even linking to it. E.P. Fischers book tells the story about an interesting man dedicated to science – who learned physics and applied sound principles to biology. “Enter without knocking” does not have any special meaning (as E.P. Fischer confirmed me) it was simply a rough-running door. Besides the biographical sketch and the detailed description of phages and phycomyces (p15) there are many moments in time that I really liked very much.
/-/
Hershey (with whom he shared the Nobel in 1969) should give a lecture and asked about the background of the audience. Max Delbrück answered by a postcard 6/1/1943: think of “complete ignorance and infinite intelligence” – the lecture became a success.
/-/
Delbrück always emphasized (p 148) that data should only be augmented by those who can put old data into new hypotheses. He even said “enough data” as thinking about current experiments is being as important as doing new ones. Both, thinking and doing experiments, should even be more important than publishing (his lifetime list has 115 items). Writing up results should serve as a method to connect what is currently known and what will be known. Delbrück even suggested to spend – “one day per week without pipettes”.
/-/
From an interview (p 240): “Genetic engeneering may possibly be a large thread for the future but possible also the biggest hope”.
/-/
Delbrück was a great admirer of Eliot, Rilke and of Beckett. Samuel Beckett in “Waiting for Godot” probably inspired “Licht und Leben” (p 260): “We give birth astride a grave, the light gleams an instant, then there is night once more.”
Addendum
Page 239 contains a mystery: In 1978 Delbrück gave a lecture at Caltech where he wanted to include a citation from Kierkegaard: “Wissen ist eine Sache der Einstellung, eine Leidenschaft, eigentlich eine unerlaubte Einstellung. Denn der Zwang zum Wissen ist wie Trunksucht, wie Liebesverlangen, wie Mordlust, in dem sie einen Charakter aus dem Gleichgewicht wirft. Es stimmt doch gar nicht, daß der Wissenschaftler hinter der Wahrheit her ist. Sie ist hinter ihm her. Er leidet unter ihr.” Delbrück, however, could not find the correct source, even announced to pay 50$ for it. I also looked at my library but couldn´t locate it – who knows the source?
More -omics
Genomeweb Daily News has a short -omics story
describing the 2-year-old project as one that will “have more immediate impact on medicine and medical practices than the Human Genome Project,†the University of Alberta said researchers have catalogued “2,500 metabolites, 1,200 drugs, and 3,500 food components that can be found in the human body.â€
Here is the the Human Metabolome Database by Genome Alberta that reports slightly different figures on the intro page (attn HMDB is not identical to the Human Mitochondrial Database!)
The database is designed to contain or link three kinds of data: 1) chemical data, 2) clinical data, and 3) molecular biology/biochemistry data. The database currently contains more than 2100 metabolite entries including both water-soluble and lipid soluble metabolites as well as metabolites that would be regarded as either abundant (>1 mM) or relatively rare (<1 nM). Additionally, approximately 5500 protein (and DNA) sequences are linked to these metabolite entries.
The NAR Jan issue has the accompanying paper:
Metabolomics is a newly emerging field of ‘omics’ research concerned with the high-throughput identification, quantification and characterization of the small molecule metabolites in the metabolome.
(Re)programming adult into pluripotent cells
Last August I found an interesting paper in Cell that could mark a scientific breakthrough. In a step down approach the authors were able to reduce a mix of 24 transcription factors to 4 that were still able to induce a mouse embryonic stem cell signature (by using a fusion cassette of ß-galactosidase and neomycin resistance into Fbx15 gene).
The magic cocktail consisted of Oct3/4 and Sox2 (both embryonic stem cell core factors that directly target Fbx15), c-My (does global histone acetylation) and Klf4 (represses p53 directly).
My first question is if this cocktail reprogramms differentiated cells or if it just selects rare progenitors otherwise hidden under more fibroblast cells. My second question is if these are fully reprogrammed cells – or if the Fbx15 signature is somewhat misleading. My third question: Is this effect mouse specific?
I have now checked ISI if any paper is already citing this work – it seems that we need more patience. Yea, yea.