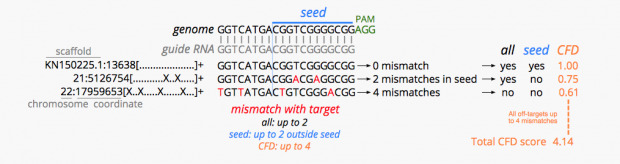
Unfortunately we don’t know the genome of the Chinese parents which would be necessary for any prediction of genome mutations in the twins . So far we only know one confirmed off target site from Dr. He’s Hongkong slides that reports 5 mismatches.

At least I would expect for statistical reasons, some 1 bp mismatch, some 2bp, some 3bp…, but not just one 5bp off target site.
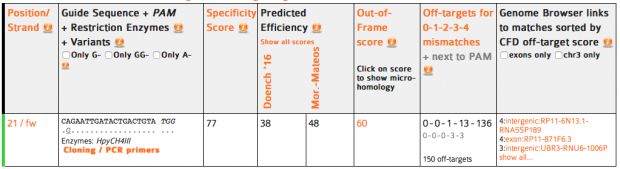
So we could check several online services with the know sgRNA sequence that Dr. He used.
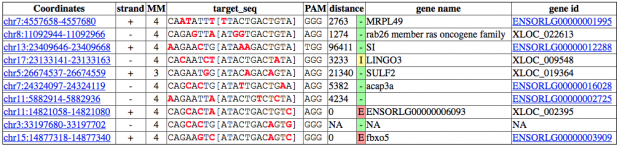
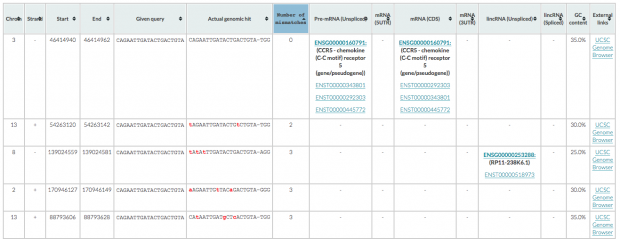
CRISPOR finds one region with 2 mismatches (intergenic) and 13 with 3 mismatches (7 intergenic hits, 5x introns (ST8SIA6, CTD-2532D12.5, CNTN5, C2D3, RABGAP1) and 1x exon of ACACA). This doesn’t match the reported off target sequence.
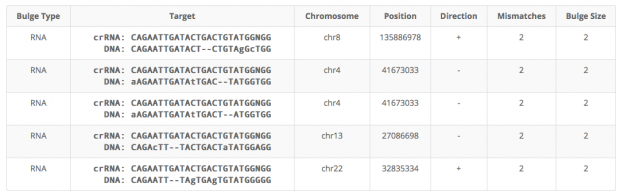
Cas-OFFinder identifies 5 regions with 2 mismatches (1x intergenic, 2x sites in LIMCH1, 1x intergenic, 1 in BPIFC).
CCtop (yes, I was waiting for that name :-)
COS finds only one intergenic region with 3 mismatches.
has the best explanation of what it does but cannot search from sgRNA to off-target as many other online tools.
is spotting 1 intergenic region with 2 mismatches, 3 with 3 mismatches (2 intergenic, 1 in RP11-238K6.1).
Taken these five different predictions together, I can’t make conclusion but think that we need to resequence the families with much higher coverage (150-200x fold coverage) and even compare the mutations with the phenotype of the children.