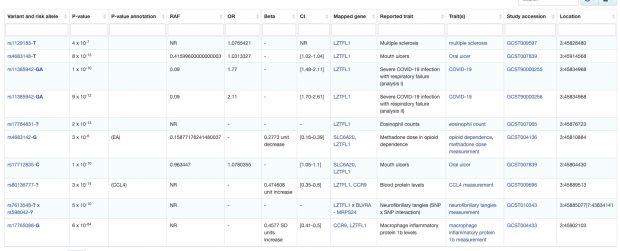
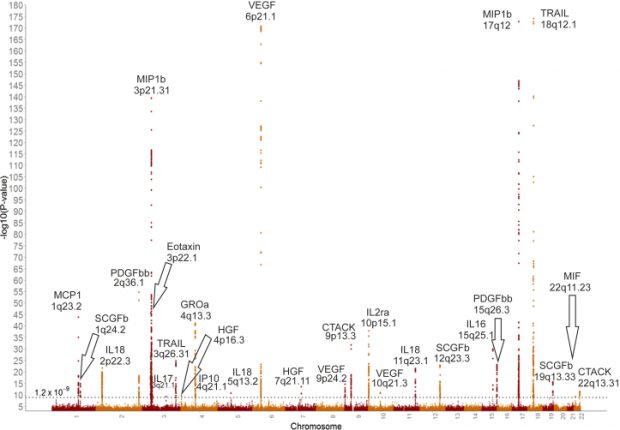
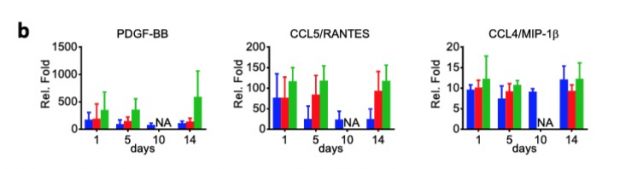
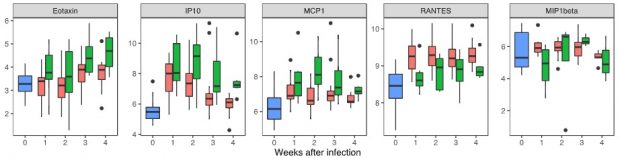
There is a great comment over at PubPeer about a rather weak and biased paper about 15 years of GWAS studies
Some of us are old enough to remember what was originally promised by genome-wide association studies (GWASs): we would finally discover the genes aetiologically involved in the conditions which till then we had been researching using a combination of linkage and candidate gene association studies. Clearly, this has not happened. With the benefit of hindsight and a myriad actual results we now clearly appreciate what perhaps we should always have realised, which is that common variants do not have substantial effects on phenotypes. GWASs yield complex, difficult to interpret findings which implicate variants but not genes and have not delivered the insights which we were promised they would.