I had the chance to install now the new PLINK GWAS software for a further analysis of recently published ORMDL3 asthma data. It seems that PLINK is some software that I was looking for a long time (paper link|download link). There are great and foolproof functions to check the validity of your data. I discovered for example unnoticed stratification in the German case-control sample by first and second component of the MDS analysis

In addition there are single unexplained outliers, see the box and whiskers of the Z scores:
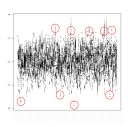
On the other hand there were even unreported sibs in this data set. Further problems were found with the lead SNP rs3894194 – showing significantly more missings in affected than unaffected individuals (p=0.007). Fortunately, however, there was no cluster heterogeneity as may have been assumed by the MDS analysis.
The most exciting part comes with the new PLINK function proxy testing. As I believe in effects of rare variants that may have not been typed (like in FLG), the standard approach by tag SNPs may be not very informative. The accompanying PLINK paper explains
… if multiple rare disease variants exist within the same gene or genomic region, then, instead of standard association, one might consider an approach more akin to linkage analysis but performed in population-based samples of unrelated individuals. Rather than directly test frequency differences of a variant, we propose examining ancestral sharing at a locus, following ideas from previous work on haplotype sharing methods.
As for some functional arguments I am mainly interested in the downstream region of the LD block I am choosing SNP rs9303277 as anchorpoint. This gives the following result:
The OR doubles! with a rare variant — while no such effect is being found in the ORMDL3 region. There are also some epistatic effects:
Does this have any biological meaning?

