You may want to read the full story at Public Rambling (fiction), the NYT article of Amy Harmon (fact) or the “Peepshow” article of Marco Evers (fact) and come back here afterwards.
The recent advances in genome sequencing (and typing) has left us with an enormous amount of data. Although technology has been available for a couple of years knowledge exploded only recently, where people now may decide to participate in a genome study or even have their genes tested on their own costs at DeCodeMe, 23andme, Navigenics or other personal genome service provider.
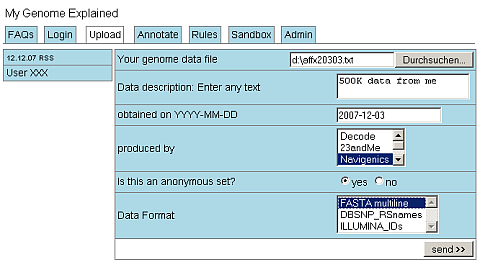
The main question is, what do these data really mean for us? Should we start an Open Source Project Genome Explained to collect the necessary annotation rules and provide a platform to apply these rules to local data? The data mode may be quite simple: a multiline FASTA flatfile where each sequence is associated with one or more disease risks.
 .
.
For individual advice, however, we need to take into account also the interaction with risk lowering sequences which is impossible. So this is not a technical question: there are so many ethical questions that need to be answered first. “Do not harm” is a longstanding principle in the medical field.
Nachtrag
Comments are closed.