I have already seen this data in Washington and even talked to one of the two first authors (pun!) at the airport – the first genomewide scan for atopic dermatitis is now being online at the nature genetics website. The overall effects are disppointing small – my quick plot gives the cumulative (sic!) negative log p values.
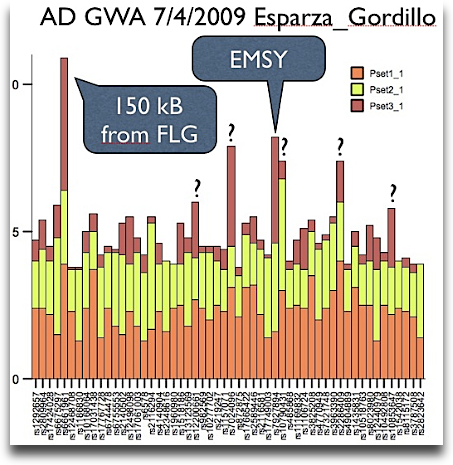
If we agree that the best signal is being caused by filaggrin, and this signal is being found, it is unlikely that there will be so much additional gain in knowledge by just sequencing the next generation.
Furthermore, it is a big mystery to me why the paper is written around the C11orf30 peak as there is no evidence for any role of EMSY in AD pathogenesis. What about the other tiny peaks? For example rs12200661 @ F13A1 / LY86, rs7024096 @ CDKN2B / DMRTA, rs2286409 @ PCNX / SIPA1L1 or rs10853647 @ HRH4/ZNF521? I think scientists should get also prices for NOT writing some kind of papers – just because the technology was there.