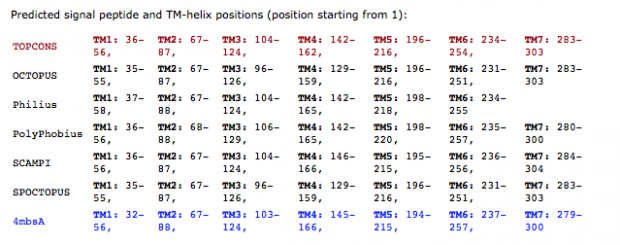
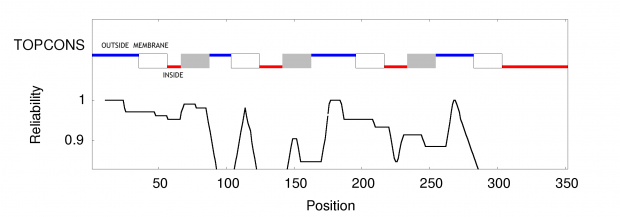
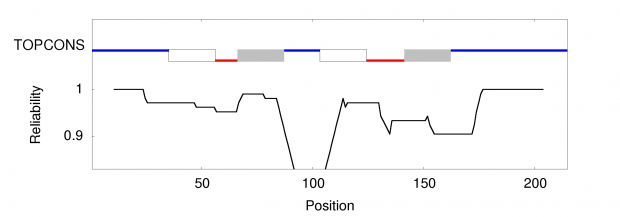
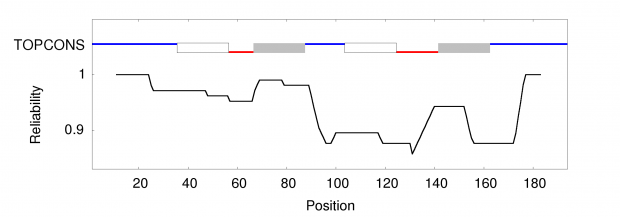
When working on a forthcoming talk about the ethics of correcting gene defects, I asked myself: Are there any empirical examples where the correction of a so called “deleterious” mutation may be a disadvantage? Or in other words: Are there any beneficial side-effects of otherwise deleterious mutations?
(Don’t answer this with the joke that the Y chromosome is a X with a large deletion :-)
Yes, there are some examples of heterozygote advantage
- HBB-p.E6V leads to sickle cell anemia and malaria resistance
- CFTR Delta F508 leads to CF and protects against tuberculosis
- …
Maybe I am not asking if there are beneficial/deleterious mutations in a single gene – the question here is more about distant / cis-regulating elements. And there seems to be a thesis that
deleterious mutations have long been thought to be unimportant, however this view overlooks the pivotal role of epistasis. The unique experiments presented here give new insights into the historical and highly contingent nature of evolution. While evolution frequently finds a well adapted solution in the long-term, evolving populations will frequently climb suboptimal peaks initially. Deleterious mutations become useful because they aide evolution in reconciling short-term and long-term adaptation.
the thesis made it also into a PNAS paper
It might seem obvious that deleterious mutations must impede evolution. However, a later mutation may interact with a deleterious predecessor, facilitating otherwise inaccessible adaptations. … We studied digital organisms—computer programs that replicate and evolve—to compare adaptation in populations where deleterious mutations were disallowed with unrestricted controls. Control populations achieved higher fitness values because some deleterious mutations acted as stepping stones across otherwise impassable fitness valleys. Deleterious mutations can thus sometimes play a constructive role in adaptive evolution.
Looks like humans shouldn’t interfere with their own evolution as long as the rules are not known… The PNAS paper above has been cited many times, it will take some time to scan these for more empirical examples.