Wafik S El Deiry has now uploaded more than 40 times a rebuttal letter to PubPeer complaining about being bullied by “academic terrorism”. Thomas C Südhoff is even more aggressive with new ad hominem attacks as I just learned

while his explanation of the numerous duplications is clearly wrong
… the tiny allegedly cloned areas of similar background signals partly overlap and are randomly distributed in the image. Besides the fact that it would make no sense to duplicate such small areas of background – a fraudster could just run a gel with empty lanes – and that such duplications do not improve the data, overlapping duplications like this are nearly impossible to manufacture.
Of course also small areas can be copied with the clone tool. If the placement is random or intentional can only be judged from the original image while an educated guess is certainly allowed. Running Photoshop is at least far more time and cost effective than running a gel with an empty lane.
A general problem here is that digital reproductions of images – both of immunoblots and of tissue sections or cells – can create artifactual microduplications especially if the image resolution is changed during reproductions.
This is outright wrong. Artifacts by capturing or stitching software is possible in theory while in practice we have found it only a few times.
So here comes my assessment of the now famous Synaptotagmin-1/Synaptopyhsin-1 immunoblot
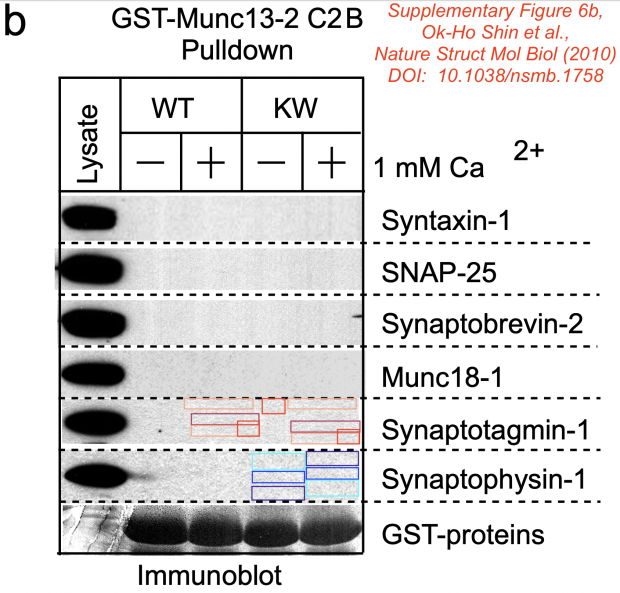
The assessment is based on the directly extracted (inline) image from the PDF.

Identical patches were confirmed using 3 software packages Forensically, ImageDup and ICMF.

Also the manual annotation below shows 100% identical areas where the KW+ lane pixel has been copied to KW- (the other direction is less likely). Not sure what had been there, dust, dirt, text marker or another dot?

Südhof comments on this image on his website
Mistake identified: Dr. E. Bik claims that the Suppl. Figure 6b immunoblot stripes (reproduced digitally at low resolution by the journal from a non-digital original blot) contains tiny areas of microduplications in the background pattern (not the actual signal). These areas are tiny, within a blot, randomly distributed, and only digitally identifiable. She implies that these blots are suspicious and could be manipulated.
Resolution: This is an unusually bizarre accusation since it refers to digital low resolution images in which tiny image areas would have been scrambled by a person if Dr. Bik’s accusation were correct. Even though she maintains publicly that she won’t speculate about motivations, her accusations imply a motivation that would be difficult to understand since any manipulation here would produce a partly altered background. The most likely explanation here is, like for many of the ‘mistakes’ identified by Dr. Bik’s A.I.-powered software, that these random microduplications are simply a reproduction artifact of a digitized image.
Classification: unfounded
Great story: The journal Nature Structural & Molecular Biology received the original blots and digitized them? So this is their fault? These are neither tiny spots, nor are they randomly distributed and of course, they can be seen by naked eye.
German newspapers covered the Südhof stor already (SPIEGEL, FAZ but also Science Magazine). Ulrich Dirnagel/Tagesspiegel believes that any intentional manipulation or deception cannot be recognized. I am not sure when looking at the images above.