When starting again analysis of pooled DNA samples – this time on the Affymetrix platform – I discovered not only the much improved “Genotyping Console” but also that my earlier Genepool binaries do not work anymore under VMWARE/Ubuntu. I therefore tried Continue reading My linux box
Tag Archives: Affymetrix
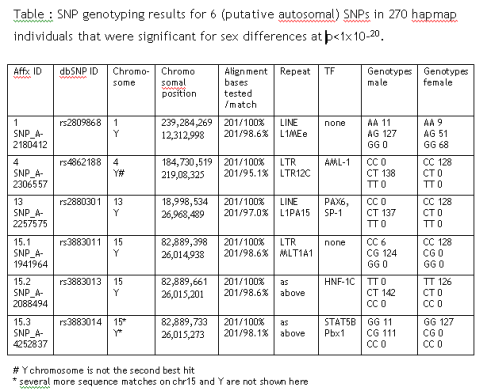
Autosomal inheritance of sex-linked marker
While optimizing the analysis strategy for a 500,000 SNP Affymetrix array set, I found 6 autosomal SNPs that show highly significant sex-dependent allele differences: rs2809868, rs4862188, rs2880301, rs3883011, rs3883013 and rs3883014.
Sure, there could be autosomal marker that influences male/female outcome but there is a more likely explanation: All SNPs have paralogue sequence stretches on the Y chromosome that are co-amplified during PCR. From the initial genotyping results it is most likely that only the Y chromosomal stretch is being mutated in SNP 4, 13 and 15.2.
These SNPs are perfect sex marker, as they include an autosomal control allele (in comparison to pure Y markers like SNPs in SRY). They are always unambiguous (in contrast to pure X marker where only heterocygotes are informative).
They even offer advantage to commercial STR kits of the Amelogenin/Amely gene situated (in the Y parautosomal region) as they would not be affected by excess homologous X chromosomal material as often found in forensic situations. In addition, they might overcome some other weakness of the Amelogenin test where a second assay is usually recommended.
If you will ever see a case-control study that is highlighting any of these SNPs, you can be sure that this study had a distorted male-female ratio between case and controls.
Pathway to nowhere
I love pathway diagrams since I mounted the famous Biochemical Pathways of Boehringer in my bachelor flat. As far as complex disease genetics is concerned with many disease genes, an integration into a pathway context becomes critical. There are many attempts to extract this information from the literature and many companies that offer highly curated information (Biomax, Ariadne Genomics, Genomatix to name a few). Academia relies mainly on KEGG, the Kyoto encyclopedia of genes and genomes or Biocarta. Last week another pathway server appeared that is curated by the NCI and Nature magazine. Let’s have a look – I am currently working on Affymetrix 500K SNP annotation, yea, yea.