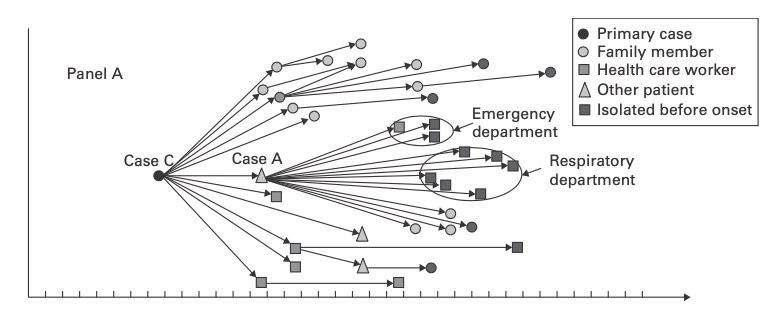
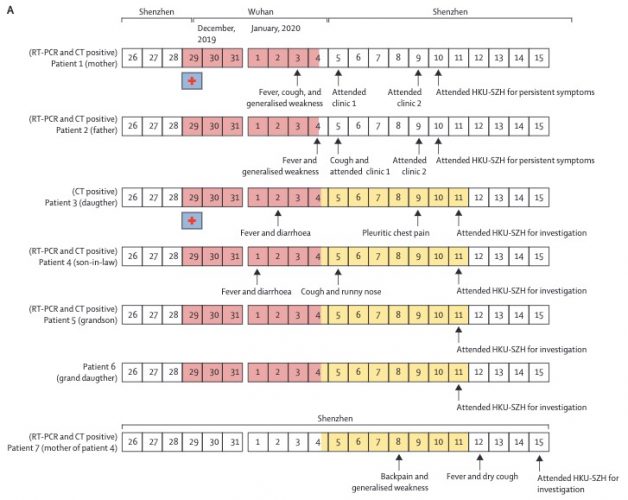
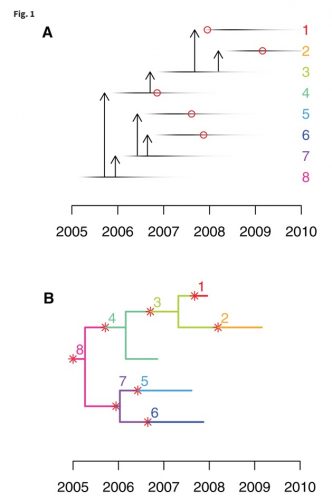
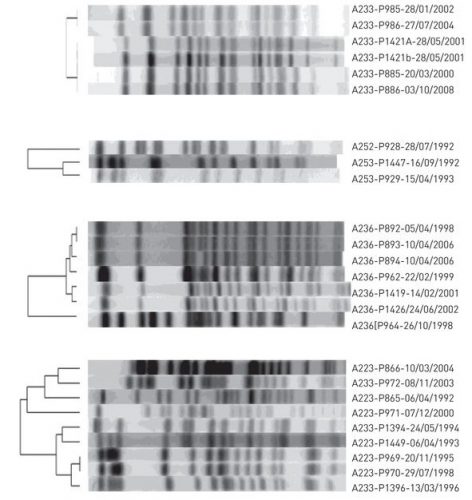
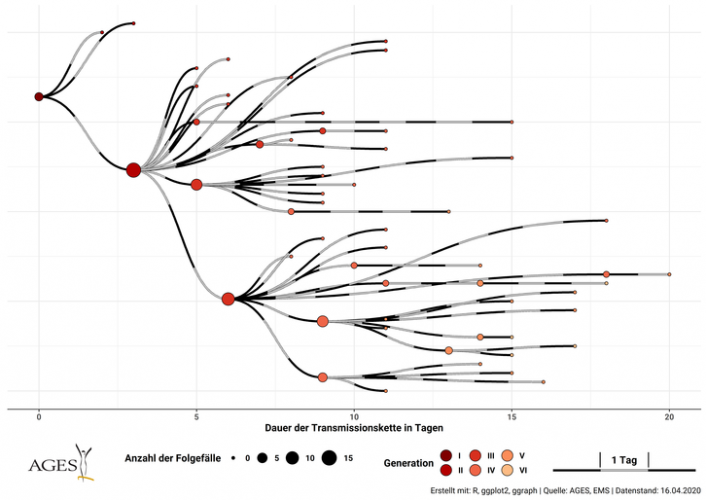
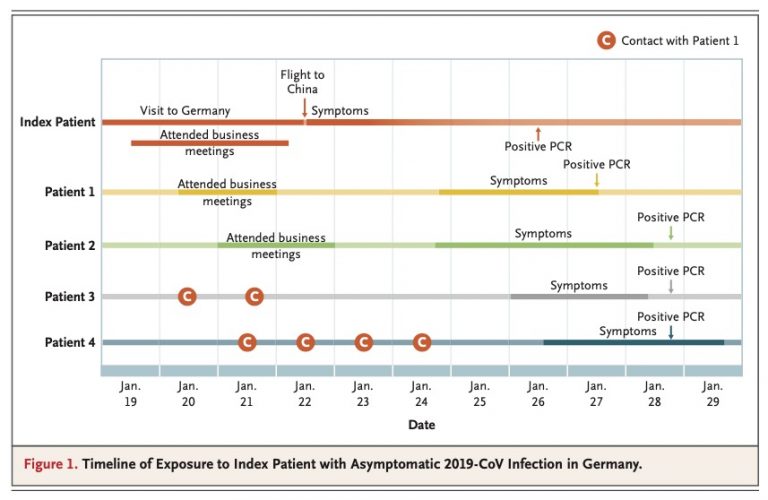
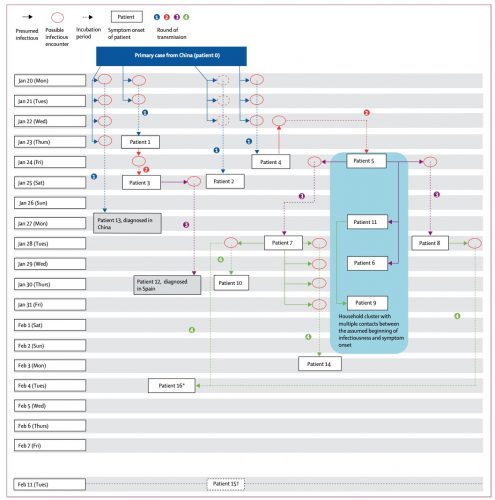
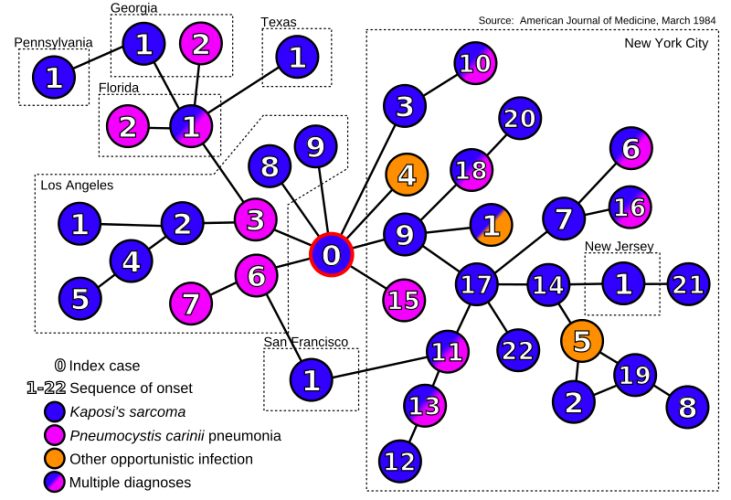
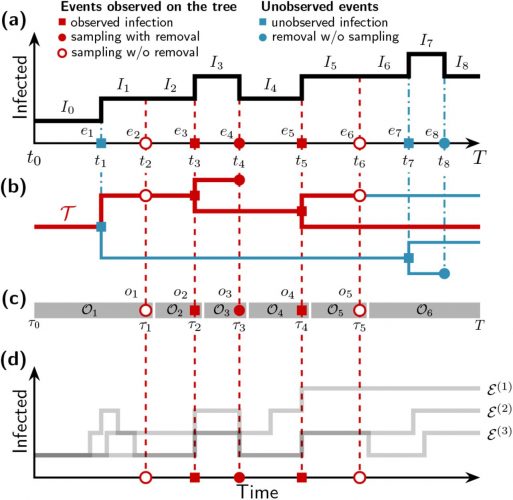
There was the 2077 Goh PNAS paper using that title. And it is a sound approach probably better than any division of chapters in Harrison’s Internal Medicine!
A network of disorders and disease genes linked by known disorder–gene associations offers a platform to explore in a single graph-theoretic framework all known phenotype and disease gene associations, indicating the common genetic origin of many diseases. Genes associated with similar disorders show both higher likelihood of physical interactions between their products and higher expression profiling similarity for their transcripts, supporting the existence of distinct disease-specific functional modules. We find that essential human genes are likely to encode hub proteins and are expressed widely in most tissues.
I found this on a slide at the recent vitamin D congress in London and was just interested to see, how often this paper has been cited. So far as I remember only the Barabasi update. And the result is impressing Continue reading The human disease network (no need to call it diseasome) →