- I assume that your digicam also produces a .mov file
- Install avisysynth and virtualdubmod
- create a file bridge.avs with the only line
DirectShowSource (“c:\video\videofile.mov”)
- Open bridge.avs in virtualdubmod, edit the video as usual and save it in .avi format
- Convert the .avi file into flash .flv as described earlier
Tag Archives: Uncategorized
NHANES R data parser
NHANES is a great ressource for doing epidemiological research. As the NIH website provides only data import for commercial software here is my rewrite in R. First load from their site
adult.exe
youth.exe
lab.exe
lab2.exe
exam.exe
put everything in one directory and expand the self-extracting archives. Then create from each SAS file a new variable content file that will only contain variable name and tab separated start position in the .dat file. Adult.var for example would read like this:
SEQN 1
DMPFSEQ 6
DMPSTAT 11
DMARETHN 12
DMARACER 13
...
HAZNOK5R 3345
Then start the following R job with the datasets and variables that you are interested in
1: | |
Quick database edit from webbrowser
Frequently, I need to change only one single field in a database. For that purpose I am using a short php script – it basically outputs all fields and highlights them with a javascript popup where you can do any changes (or even delete a row) while a “return” writes the change back to the database.
You will immediately see, where the script needs some adaptation – all the trick is in line 28.
line 2: your database
line 5,9…: your mytable
line 23: your columns
1: | |
R parallel computing
Following several unsuccessful attempts to implement a parallel computing platform for R statistical software, I am showing here my current approach that is largely influenced by a recent paper on cluster programming in c’t 6/06 by Oliver Lau (sorry, no online version). My primary interest is with the R library snow (or snow-ft) that offers the function clusterApplyLB. This function is all I need for my R programs.
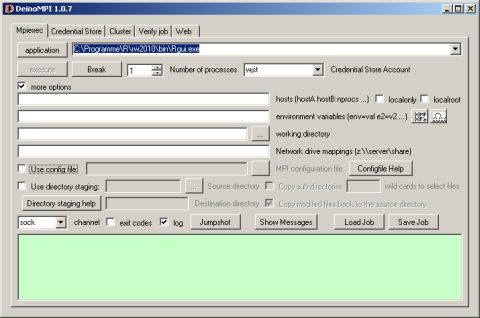
Now it gets more complicated: library(snow) depends on library(Rmpi): Hao Yu has an excellent description at www.stats.uwo.ca/faculty/yu/Rmpi how to set up the mpi layer with MPICH2. I am currently experimenting with DeinoMPI a closely related high performance Windows interface. According to its developer David Ashton it has the following advantages
First, DeinoMPI does not require MPI applications to be started by mpiexec in order to call MPI_Comm_spawn so you could load Rmpi from the Rgui.exe without having to bother with calling mpiexec. Second, DeinoMPI loads the user profile when starting applications so if you query the user’s temporary directory you will get the user specific path and not the Windows system temp directory. Third, DeinoMPI handles arguments with spaces correctly if you quote them so you can pass environment variables with spaces in them. Fourth, DeinoMPI allows you to use the MPI Info object to pass extra options to MPI_Comm_spawn like drive mappings. So you could create an MPI_Info object and set wdir=z:\ and map=z:\\server\share. Then pass this info object in with the MPI_Comm_spawn command and you could map a network drive and launch an executable from this drive.
So far the Rmpi package is compiled for MPICH2 (not DeinoMPI) so it won’t run with only DeinoMPI installed but there is a good chance that this will change in the near future.
Further useful references are in the R newsletter 2003, p21 cran.r-project.org/doc/Rnewsand a paper in the UW Biostatistics Working Paper Series on “Simple Parallel Statistical Computing in R” by Anthony Rossini and LukeTierney.
BTW, haplotypes of the hapmap project were computed on a 110 node cluster provided by both Peter Donnelly’s Mathematical Genetics Group www.stats.ox.ac.uk based at the Oxford Centre for Gene Function and by a 128 node compute cluster provided by the Oxford e-Science Centre e-science.ox.ac.uk as part of the National Grid Service[to be cont’d…].
Geo IP identification
The Geo IP database is available at Maxmind and allows to trace your home city from IP addresses. Here is a quick and dirty script to upload the Geo IP data into MySQL:
1: | |
I would also put an index on loc_id. Finally the database should be available as
SELECT city
FROM GeoLiteCity INNER JOIN GeoLiteCityBlocks ON GeoLiteCity.locID = GeoLiteCityBlocks.locID
WHERE $myIP >= startIpNum AND $myIP <= endIpNum;
where $myIP is calculated as
substr($_SERVER['REMOTE_ADDR'],0,3) * 16777216 +
substr($_SERVER['REMOTE_ADDR'],4,3) * 65536 +
substr($_SERVER['REMOTE_ADDR'],8,3) * 256 +
substr($_SERVER['REMOTE_ADDR'],12,3)
WordPress as CMS
I have read many useful (and also some less useful) comments how to squeeze WordPress to work as a CMS.
I did not want to make any major changes to scripts that would be lost after an upgrade. I did not want to have extra plugins to change home (for example by creating an overriding home.cfm). I did not want to have any new categories. I did not want to change permalink structure. I still need my directory plugin to work, I still need the blog (some redirects even loose the blog address!) and I wanted to keep the RSS feed.
After several hours I came up with an very simple solution: Take a standard page and rename its title and slug to “Home” – assign a special “Home” template – redirect htaccess to this page. The only trick is to make the “Home” template work: it is basically a copy of the index.cfm in your WordPress theme directory where the line calling loop.php is being replaced with a slightly modifed loop code.
1: | |
A simple POP answering machine
A script that I used for many years…
1: | |
A low-cost system for a PDF literature archiv II
With Google Desktop you will have the archiving capabilities that would have cost 10,000$ only a few years ago. Things become more tricky if want to share your archive on a workgroup level. Here is an idea that I have found in the German Laborjournal
- Shut down your firewall
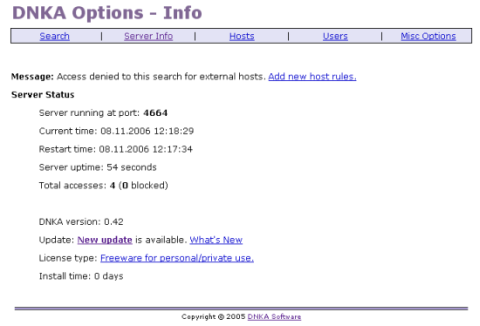
- Install the freeware DNKA available from dnka.com. It will act as a web server by interacting as a layer between Google Desktop Search and the user
- use http://127.0.0.1:4664 to configure your webserver and offer http://yourIP to your working group.
Need more information? Check geekzone and a nice indexer interface at TweakGDS.
Snapshot of your working directory
We need two open source programs: Gzip is a compression utility with a high compression rate and free from patented algorithms. GnuPG is a complete and free encryption solution to protect confidential communication and digitally stored information. Create a /backup directory in each of your working directories.
1: | |
Write the above code in a file, put it somewhere in your path and assign an icon (I am running this from the buttonbar of TotalCommander (R)). One click – and your current directory is being saved as zip file and signed with your key.
A database driven epidemiological questionnaire
During a past project I moved from a paper-based questionnaire directly to a database version. This has the advantage of generating at the same time an online version (with some interface scripts) and an offline version (by using a serial paper printing).
The whole package is written in standard HTML with some Cold Fusion(R) extension (where you need the Railo(R) or Blue Dragon(R) interpreter too).
Download (md5:1922125540), create a datasource “aerzte” and attached the included Access(R) database. Then run the following script in your browser.
1: | |
Convert external maps to OziExplorer
Many map software vendors like German TOP 50 do not include export functions. Printing, however, is usually not a problem.
I therefore suggest to use the print function for exporting map data. Just print your desired map area into a PDF file (TOP 50 nicely allows to set anchor points for that).
A PDF printer driver will be already installed on many systems (if not, please go to sourceforge and download pdfcreator). Write down the upper left and lower right GPS coordinates of your rectangle as you will need them later in OziExplorer.
You will need to download also two graphics packages: netpbm and xpdf.
Move your PDF print export and all downloaded executables into one directory. After running the following script you will see a bmp file that can be imported and calibrated in OziExplorer. Happy navigating!
1: | |