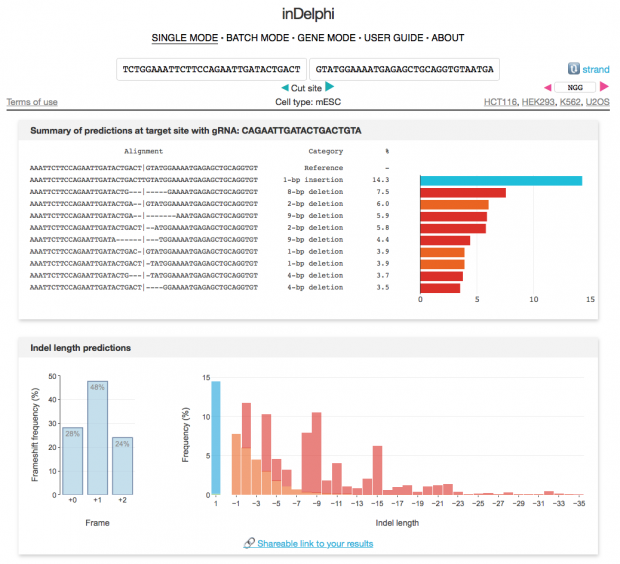
Using the new inDelphi (Nature 2018) prediction we can examine the gRNA guided cut used for the CRISPR Baby experiment. The results are somewhat unexpected
Maybe it is difficult to extrapolate from mouse to human embryonic stem cells but one observed event is not even listed here.
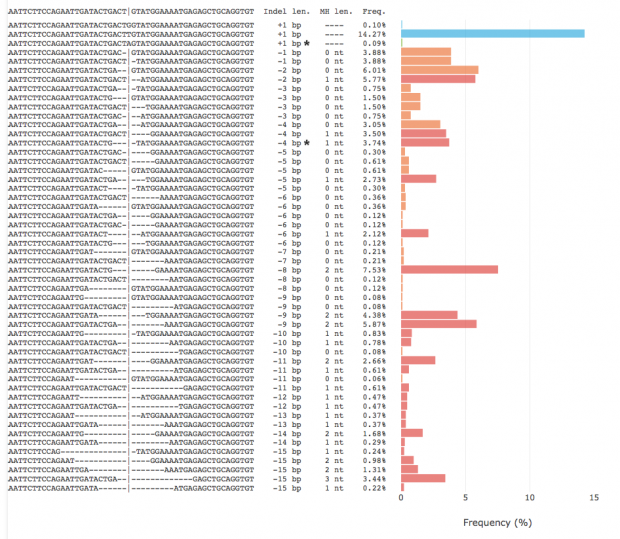
The -15 genotype has a probability of less than 0.05%. For +1 genotype the probability is 0.09% and for the -4 deletion it is 3.74%.
Looking therefore again at the Hong Kong slides of He Jiankui, I am getting doubts if the chromatogram of embryo 2 is correct interpreted even if we admit that the labels of embryo 1 and 2 have been switched..
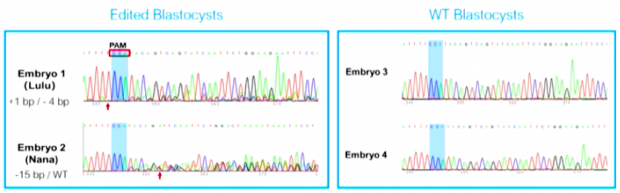
Embryo 2 does not show a clean sequence at all and certainly not a -4/+1 genotype as indicated. The sequence “ATTTTCCATACAG-ATTCAATTCTGGACTAAAATAAATACCT” isn’t even a human sequence at all.