 The Isaac Newton Institute for Mathematical Sciences has some interesting videos online – no need to travel – just sit back and relax, yea, yea.
The Isaac Newton Institute for Mathematical Sciences has some interesting videos online – no need to travel – just sit back and relax, yea, yea.
“Science advances in stages and no story is complete”
Sometimes we think we are alone with our difficulty of getting a good paper published. In lucid moments, however, we find others making the same experience- see a commentary in Science
The stress associated with publishing experimental results–a process that can take as long as obtaining the results in the first place–can drain much of the joy from practicing science Continue reading “Science advances in stages and no story is complete”
Accurate reporting
One of the PLoS editors has a vocal report on a recent meeting “Why accurate reporting is an ethical duty“. When dealing here with a misconduct case, I had the impression that many colleagues as well as some other editors think of Continue reading Accurate reporting
I am all that hath been, and is, and shall be
Following the vision of an universal god in the recent post, Assmann traces also Cudworth’s invisible god who himself cites Plutarch. From “On Isis and Osiris”: Continue reading I am all that hath been, and is, and shall be
dum eadem res intelligatur
Reading now Jan Assmann’s brilliant “Moses the Egyptian” I found Varro quoted (116-27 BC) –
Nihil interesse censens quo nomine nuncupetur, dum eadem res intelligatur
names may be of less interest as long as the “right things” are being meant.
The book was recommended to me by a guide in the new Munich synagogue as an unorthodox view of Moses and the origin of monotheism. Continue reading dum eadem res intelligatur
Fabrics for atopic dermatitis
Most of the medical research into causes of allergy is more or less irrelevant to allergy sufferers. Here is an exception [author] if you are suffering from atopic dermatitis [paper] Continue reading Fabrics for atopic dermatitis
Webcam based laser point tracking
If you do have the same problem like me – operating a computer, pointing towards a wall and looking at the same time at an audience is somewhat odd – here may be a solution for you.
Initially found this at a blog with some java source code.
Another video example is
with abandonded C# code that compiled nicely on my mac (laserinteraction). Experiments are ongoing…
Probability that a genetic association is false positive
An (anonymous) reviewer of our forthcoming EJHG paper on IgE and STAT3 pointed me towards a JNCI paper that has a nice supplement – an excel sheet to calculate the probability that a positive report is false. It basically relies on (i) the magnitude of the p-value (ii) statistical power and (iii) fraction of tested hypothesis. While we certainly know (i) and (ii), (iii) is always hard to know with many datasets including hundreds of traits that allow indefinite numbers of subgroups. Are you really interested in a new paper about “An African-specific functional polymorphism in KCNMB1 shows sex-specific association with asthma severity” that encompasses 1 of virtually 100 ethnic groups; 1 or virtually 25000 genes; 1 of 2 sexes; 1 or virtually 50 asthma related traits, yea, yea.
View plink GWA results as genome browser track
This R script will parse plink‘s GC adjusted output to a GFF file that can be uploaded as genome browser track file. Just adjust the pathname at the beginning…
1: | |
Will the Data Deluge Make the Scientific Method Obsolete?
A new Edge article answers this question. According to Chris Anderson, we are at “the end of science”, that is, science as we know it.
The quest for knowledge used to begin with grand theories. Now it begins with massive amounts of data. Welcome to the Petabyte Age.
Yesterday I reviewed a paper that crunches massive amount of data (and even found a new pathway for asthma). Nevertheless I was asking the question if this wishful thinking? Just take the next gene in one region and the overnext in another one and I would come up with a completely different pathway. This is all about association and not by the traditional “theorize, model, test it” way of science we have been brought along, yea, yea.
Asthma and allergy trend
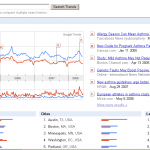 Besides traditional and expensive epidemiological studies to compare disease prevalence over time, the net offers also some nice research tools – click on the thumbnail to see Google asthma trend results by today. The query counts are largely consistent with most recent scientific papers seeing the asthma epidemic now at a (high) plateau. Did the environmental risk now saturate all individuals at risk?
Besides traditional and expensive epidemiological studies to compare disease prevalence over time, the net offers also some nice research tools – click on the thumbnail to see Google asthma trend results by today. The query counts are largely consistent with most recent scientific papers seeing the asthma epidemic now at a (high) plateau. Did the environmental risk now saturate all individuals at risk?
The best climbs
It is a fact that the best climbs are performed by famous mountaineers before they became famous (Doug Scott in “Mountain” according to an inscription at Firmian/Messner Mountain Museum). Continue reading The best climbs
Retire retirement
May 29, 2008 Nature has an interesting commentary by Peter Lawrence (66) about the archaic practice of retirement of active scientists at a determined age. It is a quite luxurious habit of “Doing what I like” while having a mostly pleasant life here on earth as a scientist, it may be a quite logical to prolong the scientific career.
German Human geneticist warns against personal genomics
Wolfram Henn, Vorsitzender der Kommission für Grundpositionen und ethische Fragen der Deutschen Gesellschaft für Humangenetik, warnt im Interview mit Technology Review (Ausgabe 07/08 […]) vor persönlichen Genomanalysen, die über das Internet angeboten werden. So bietet beispielsweise das Unternehmen 23andMe seit kurzem eine Genomanalyse für nur 999 Dollar an.
Welcome in the club! Just to let you know that 23andme has been stopped 2 days ago as reported by Spiegel magazine.
Déjà vu extended
Given my interest in strange phenomena leading to science misperception I wonder why I didn’t find this site earlier as it tells you also everything about Déjà Vu, Déjà Vécu, Déjà Visité, L’esprit de l’Escalier (comeback when it is too late), Capgras delusion (replaced friend), Fregoli delusion (same person appears in different bodies) and prosopagnosia (unable to recognize faces also known as myopia…). Yea, yea.