There is an interesting observation by Nick Brown over at Pubpeer who analysed a clinical dataset (see also my comment)
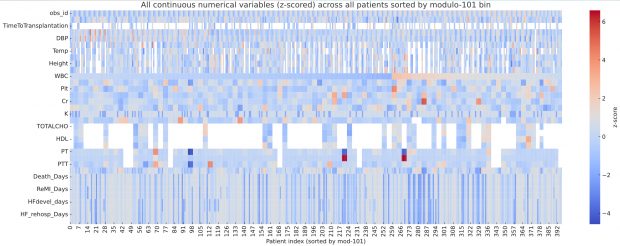
…there is a curious repeating pattern of records in the dataset. Specifically, every 101 records, in almost every case the following variables are identical: WBC, Hb, Plt, BUN, Cr, Na, BS, TOTALCHO, LDL, HDL, TG, PT, INR, PTT
which is remarkable detective work. By plotting the full dataset as a heatmap of z scores, I can confirm his observation of clusters after sorting for modulo 101 bin.
How could we have found the repetitive values without knowing the period length? Is there any formal, data-agnostic detection method?
If we even don’t know the initial sorting variable, it may makes sense to look primarily for monotonic and nearly unique variables, i.e. that are plausible ordering variables. Clearly, that’s obs_id in the BMJ dataset.
Let us first collapse all continuous variables of a row into a string forming a fingerprint. Then we compute pairwise correlations (or Euclidean distances in this case) of all fingerprints. If a dataset contains many identical or near-identical rows, we will see a multimodal distribution of correlations plus an additional big spike at 1.0 for duplicated rows. This is exactly what happens here.
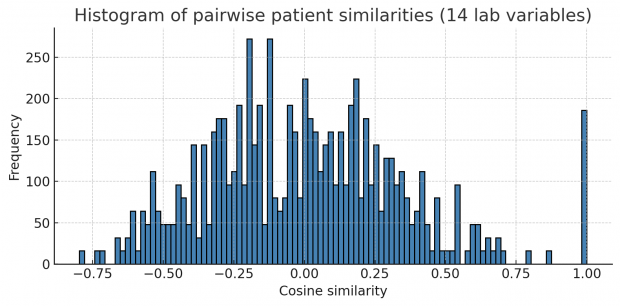
Unfortunately this works only when mainly repetitive variables are included and not too many non repetitive variables.
Next, I thought of Principal Component Analysis (PCA) as the identical blocks may create linear dependencies and the covariance matrix is becoming rank-deficient. But unfortunately results here were not very impressive – so we better stick with the cosine similarity above.
So rest assured we find an excess of identical values, but how to proceed? Duplicates spaced by a fixed lag will cause an high lag k autocorrelation in each variable. Scanning k=1…N/2 reveals spikes at the duplication lag as shown by a periodogram of row-wise similarity in the BMJ dataset.
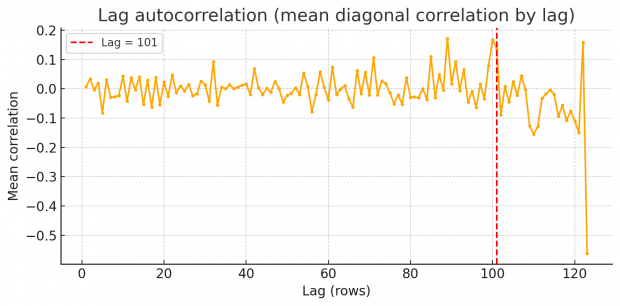
So there are peaks at around 87, 101 and 122. Unfortunately I am not an expert in time series or signal processing analysis. Can somebody else jump in here with FFT et al. ?
There may be even an easier method, using the fingerprint-gap . For every fingerprint that occurs more than once, we sort those rows by obs_id and compute the differences of obs_id between consecutive matches. Well, this shows just one dominant gap at 101 only!
We could test also all relevant mod values, lets say between 50 and 150. For each candidate we compute the across-group variance of the standardized lab-means. The result is interesting
Modulus 52: variance = 0.084019 Modulus 87: variance = 0.138662 Modulus 101: variance = 0.789720
As a cross check let us look into white blood cell counts (WBC) and hemoglobin (Hb).
I am not sure, how to interpret this. Mod 52 may reflect shorter template fragments but did not show up in the autocorrelation test. Mod 87 has rather smooth, coherent curve and is supported by autocorrelation. Mod 101 is more noisy, but gives probably the best explanation for block copying values. Maybe the authors block copied at two occasions?
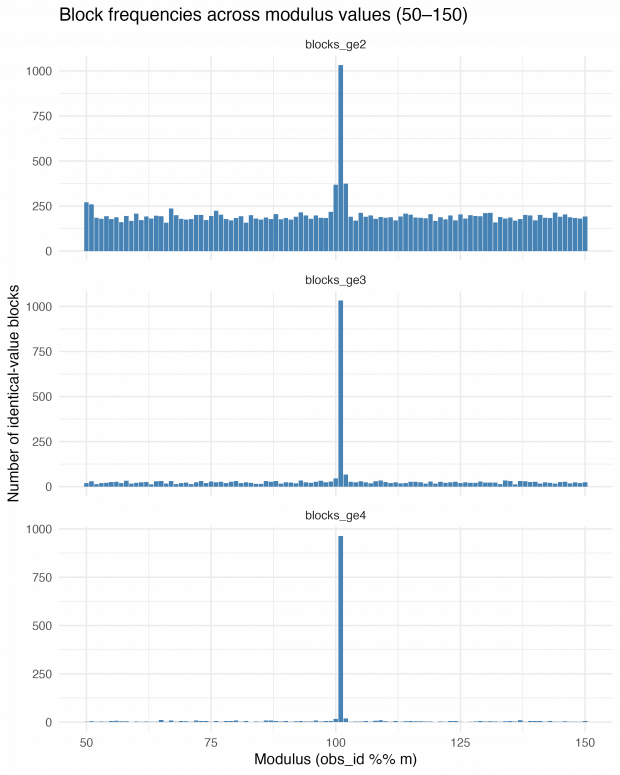
On the next day, I thought of a strategy to find the exact repetition numbers. Why not looping over mod 50 through 150 and just count the number of identical blocks? This is very informative – blocks of size 2, size 3 and 4 or greater show an exact maximum at modulus 101.