Here is a quick update on some genes of my recent asthma exome paper coming now from the 1 M exome paper published yesterday as a preprint.
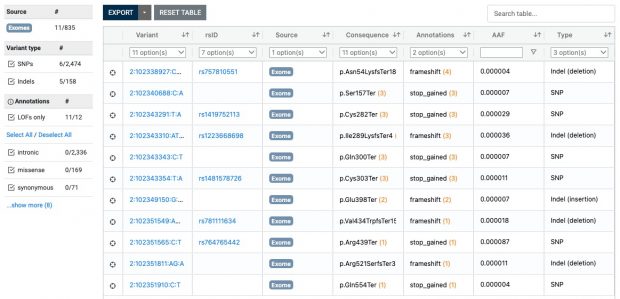
Also ClinVar shows that the IL33 receptor is not “essential” making anti IL33 receptor antibodies like etokimab, itepekimab, tozorakimab a safe therapy although not being effective in any LOF mutation carrier.
The most interesting thing in the preprint is in supplemental table 2 with the s-het values for 16,704 genes. From that table I have selected my favorite target IL33 receptor together with TLR1, ALOX15, GSDMA, IL13 and IKZF3 ( BTNL2 could not be found in the list).
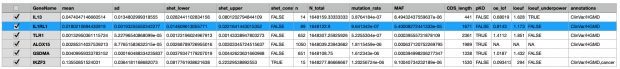
IKZF3 would be dangerous to be touched (see my 2008 commentary) while in the 2022 exome paper I also found only protective variants in the 5′-UTR but not any LOF variant – probably as IKZF3 is the only essential gene in the list.
So what’s next? I am still thinking how to reduce my exome set to the causal variants as half of the mutations are probably LD artefacts. And well, it would be super interesting to examine now two extreme inbred populations for their mutation spectrum, loosing either asthma variants by healthy (Amish) or diseased founders (Tristan da Cunha). Unfortunately there is little hope that this will happen – current science is built more on competition than collaboration.