Related posts: Parascience in nature medicine?|
Tag Archives: genetics
Genetic heritage and language does not always match
This is the interesting outcome of a new PNAS paper (pdf, R) that finds
According to the farming/language dispersal hypothesis, migrations fueled by the shift toward agriculture and animal husbandry in the Holocene have given rise to some of the largest language families identifiable today.
Most researchers thought that genes and language would match while the new paper shows
At the level of individual populations, we estimate more matches (matching enclaves or aligned populations) than mis- matches, but single-population mismatches are present in each continent and within each language family.
The database can be found at https://gelato.clld.org/
What did Sequana really find at Tristan da Cunha?
The Tristan da Cunha asthma study was leading to patent US6087485A. The patent describes
highly significant linkage in the genome scan (p=0.0001 for history of asthma and p=0.0009 for methacholine challenge) … at D11S907, a marker on the short arm of chromosome 11.
D11S907 or AFM109YA1 is a microsatellite marker located at 11p13 in a gene known as EHF (ETS homologous factor). There should be 2 genes in close proximity of the marker: ASTH1I and ASTH1J.
ASTH1I and ASTH1J were detected by exon trapping. ASTH1I exons detected a 2.8 kb mRNA expressed at high levels in trachea and prostate, and at lower levels in lung and kidney …
ASTH1J exons detected a 6.0 kb mRNA expressed at high levels in the trachea, prostate and pancreas and at lower levels in colon, small intestine, lung and stomach.
The sequences of table 2 in the patent are sufficient now to locate ASTH1I and ASTH1J. Continue reading What did Sequana really find at Tristan da Cunha?
How atopic dermatitis is linked to vitamin D and how IL33 splice variants associate to eosinophil numbers
(first published 12 Dec2020 and revised 10Dec2021)
We had a major discussion right before our 2010 paper where I argued that rare variants should have been included into our asthma/allergy/dermatitis GWAS. Ten years after there is now a nice paper using massive exome sequencing that finally includes them.
It seems that the respiratory tract isn’t so much influenced by rare gene variants but that there is a strong effect in the immune system.
And there is another interesting fact.
…Surveying the contribution of rare variants to the genetic architecture of human disease through exome sequencing of 177,882 UK Biobank participants …if we look at the …. European population who are carriers of a filaggrin (FLG) PTV, we find those carriers have significantly higher risk for well-known associations, such as dermatitis … and asthma … Concomitant increases in vitamin D levels suggest … increased sensitivity to ultraviolet B radiation.
So far, I have only assumed an asthma/allergy priming effect of oral vitamin D in the newborn gut. This paper now argues for an increased vitamin D sensitivity also in the skin of FLG dermatitis patients which is interesting given the largely contradictory data of serum vitamin D and atopic dermatitis. Maybe dermatologists should focus their research more on skin and local vitamin D turnover?
-II-
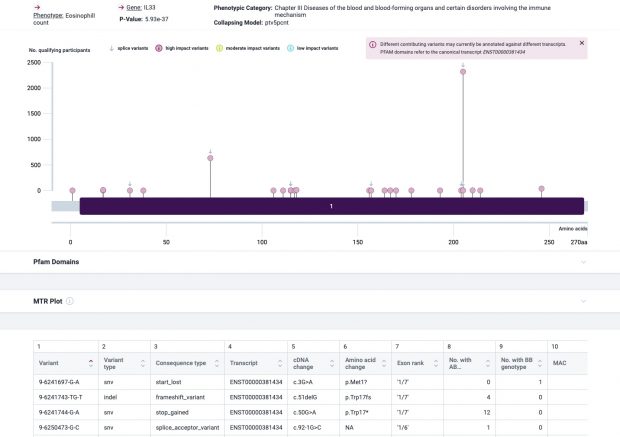
The most prominent IL33 variant carried by over 2,300 people is splice acceptor 9-6250473-G-C followed by 600+ individuals with splice donor 9-6250600-G-T.
There are not too many carriers of this variant by the sheer amount of 177,882 participants. We nevertheless know already something about the seven IL33 splice variants since 2012.
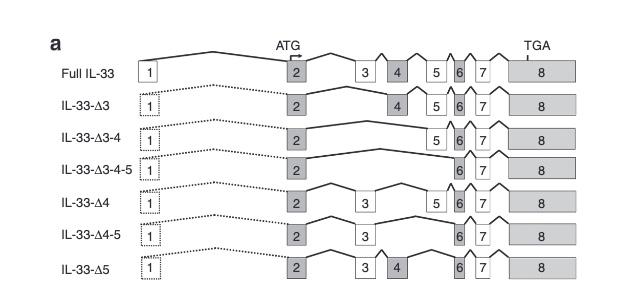
with updates in 2016
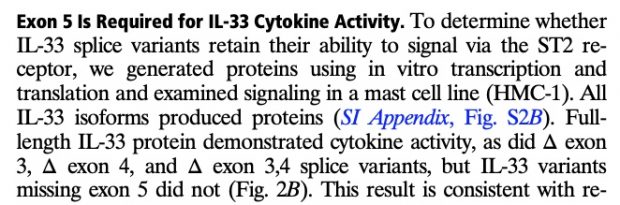
as well as in 2017
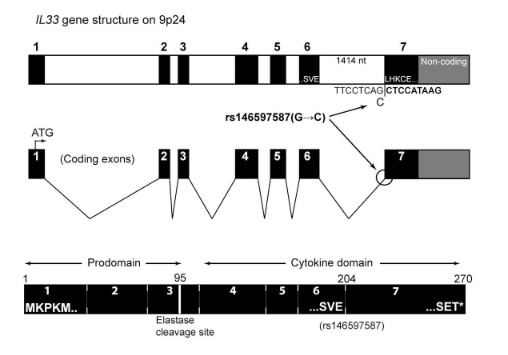
So I did a sequence match to compare the new finding with these older publications.
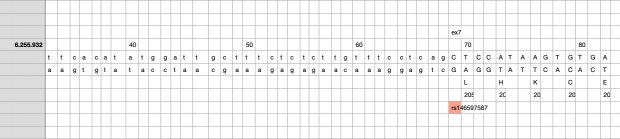
Indeed, the 2017 paper already described rs146597587 which is probably identical to the splice acceptor 9-6250473-G-C in Astra UK Phewas (genome positions do not match – I used hg19 while I don’t know the Astra reference) . Astra says also c.613-1G>C while rs146597587 is just upfront of my codon 205 (3*205=615) whatever that means.
The Astra UK Phewas at least confirms the Iceland paper above
rs146597587-C associates with lower eosinophil counts (ß= -0.21 SD, P = 2.5×10-16, N = 103,104), and reduced risk of asthma in Europeans (OR = 0.47; 95%CI: 0.32, 0.70, P = 1.8×10-4, N cases = 6,465, N controls = 302,977). Heterozygotes have about 40% lower total IL33 mRNA expression than non-carriers and allele-specific analysis based on RNA sequencing and phased genotypes shows that only 20% of the total expression is from the mutated chromosome. In half of those transcripts the mutation causes retention of the last intron, predicted to result in a premature stop codon that leads to truncation of 66 amino acids.
So it is basically a rediscovery meaning that we reached saturation.
Finally! 23 and the FDA warning
Quite some time passed already since my last post (to be exact, more than 5 years) but now there are good news. The FDA issued a warning letter on the 22nd
… The Food and Drug Administration (FDA) is sending you this letter because you are marketing the 23andMe Saliva Collection Kit and Personal Genome Service (PGS) without marketing clearance or approval in violation of the Federal Food, Drug and Cosmetic Act (the FD&C Act) … However, even after these many interactions with 23andMe, we still do not have any assurance that the firm has analytically or clinically validated the PGS for its intended uses … Therefore, 23andMe must immediately discontinue marketing the PGS until such time as it receives FDA marketing authorization for the device …
The response is quite flimsy. Yes, there may be negative side effects of genetic testing and of course tests need to validated first. Slate may be correct that the FDA’s battle with 23andMe won’t mean anything in the long run but now at least, we are set back to science, yea, yea.
Keep secret
There is a new Edge Special Event about the Hillis’s question “WHO GETS TO KEEP SECRETS?”
The question of secrecy in the information age is clearly a deep social (and mathematical) problem, and well worth paying attention to.
When does my right to privacy trump your need for security?; Should a democratic government be allowed to practice secret diplomacy? Would we rather live in a world with guaranteed privacy or a world in which there are no secrets? If the answer is somewhere in between, how do we draw the line?
With all the wikileaks hype over the last year, the Edge essay is la perfect supplement to our last paper about anonymity in genetics – check out BMC Ethics “Caught you: Threats to confidentiality due to the public release of large-scale genetic data sets“.
What we didn’t mention in this paper are more complicated statistics like stochastic record linkage – more on that in RJournal 2/2010, p.61 ff
Of the three main activities involved in scientific research, thinking, talking, and doing, I much prefer the last
is my favorite quote of a scientist – the late Frederick Sanger. I would so much love to get an interview with him.
PLINK: Bug or feature?
I am struggling now for 4 weeks with some unusual behaviour in PLINK that gives me different results with a trait of the alternate phenotype files either by calling that trait directly
plink –file mydata –tdt –pheno pheno2.txt –mpheno 1
or from a loop over all traits
plink –file mydata –tdt –pheno pheno2.txt –all-pheno
It seems that I am working with different numbers at both occasions – click to enlarge the log Continue reading PLINK: Bug or feature?
Evolutionary legacy
It’s an exceptional good science book – Cancer, The evolutionary legacy – by Mel Greaves. Having written last year a grant application about resequencing of lung specimens (and more recently a correspondence letter about the lung cancer genome that updates our earlier 31 events to 22,910) Continue reading Evolutionary legacy
Factory science
There seems to be a GWAS repository that has an entry of an asthma study otherwise not known in the biomedical literature and – as see on the screenshot -results tables are empty. Continue reading Factory science
Genetics proposes, epigenetics disposes, environment exposes
Genetics proposes,
epigenetics disposes,
environment exposes,
the human composes,
and doctor diagnoses.
(c) for the first part is by the Medawar & Medawar in “Aristotle to Zoos“.
Genome Browser now with GADview track
Just reveived an email from the creators of the Genetic Association Database (GAD)
… Your published genetic association study has been included in the recent update of the NIH based, Genetic Association Database (GAD), the database of human genetic association studies. Continue reading Genome Browser now with GADview track
Want to work with you
Over and over I am flooded with emails like
Let me introduce myself to you. I am xxxxxxxxxx, completed M. Sc Micro Biology. At present I am working as a research Fellow in Centre for xxxxxxxx, xxxxxxxxxx, India. How are you sir? I am your student. How can I mean, in January 2005 you come to India. At that time your engaged some class to us in xxxxxxxxx College, Axxxxxxxx. Presently I am working on Genetics of “xxxxxxxxxxxx†under the esteemed guidance of Dr. xxxxxxxxxxx and Dr. xxxxxxxxxx. I am very much interested to do PhD. Herewith, I am sending my curriculum Vitae as attachment to your kind perusal. I assure you, I shall work with at most devotion and sincerity to give you satisfaction and also I am confident that I can lead PhD successfully with the experience I gained during my research work at xxxxxxxxx. Given a chance I will prove my caliber.
Gene lists by automatic literature extraction
Just found at the HUM MOLGEN bulletin board a link to Fable, a new automated literature extraction system. Fable is pretty fast and can output gene lists. Sure, the screenshot below shows only those genes that I mentioned in the abstract, but this is not so bad as the most important genes wil be placed there.
BTW, the number of reviews on asthma genetics have been falling to less than 50% after closing the Asthma Gene Database. Maybe this new service will help to re-establish the former output of reviews ;-) yea, yea.
On the “Self”
If I would ever find the time, I would write a book on the “self”. Inspired by the Eccles/Popper book that I bought as a student, I always wondered how different the self is being defined in sociology, psychology/psychiatry, philosophy and theology.
As my current focus is more on genetics and immunology, I found a paper by Francisco Borrego on the “missing self” quite interesting as it highlights the genetic self is determined mainly by MHC class I molecules, where only NK cells transfected with H-2Dd were able to confer resistance for being self-attacked. It would be nice if other disciplines could also provide such simple answers, yea, yea.
Addendum
I have another suggestion: Zfp608 protects mouse mothers against immune-mediated attack by fetal cells.
Is there also a “digiself“?
Our identity has, for many years, existed quite independent of our physical incarnation in government, financial and other institutional databases. We are not real to the bank or other authorities unless we can produce something that links our physical self to our “real identity” in their database. We have many versions of this digital identity – or digiSelf, as I like to call it – spread among many databases, each with its unique characteristics, and inferred behaviours. Each one is more real to the institution – and ironically, to the people in that institution – than our physical self, what we consider to be our real self.