Materialsammlung bioinformatics / big data / deep learning / AI
Passend dazu auch der CCC Vortrag Nadja Geisler / Benjamin Hättasch am 28.12.2019
Deep Learning ist von einem Dead End zur ultimativen Lösung aller Machine Learning Probleme geworden. Die Sinnhaftigkeit und die Qualität der Lösung scheinen dabei jedoch immer mehr vom Buzzword Bingo verschluckt zu werden.
Ist es sinnvoll, weiterhin auf alle Probleme Deep Learning zu werfen? Wie gut ist sind diese Ansätze wirklich? Was könnte alles passieren, wenn wir so weiter machen? Und können diese Ansätze uns helfen, nachhaltiger zu leben? Oder befeuern sie die Erwärmung des Planetens nur weiter?
Dazu der gigantische Energieverbrauch durch die Rechenleistung.
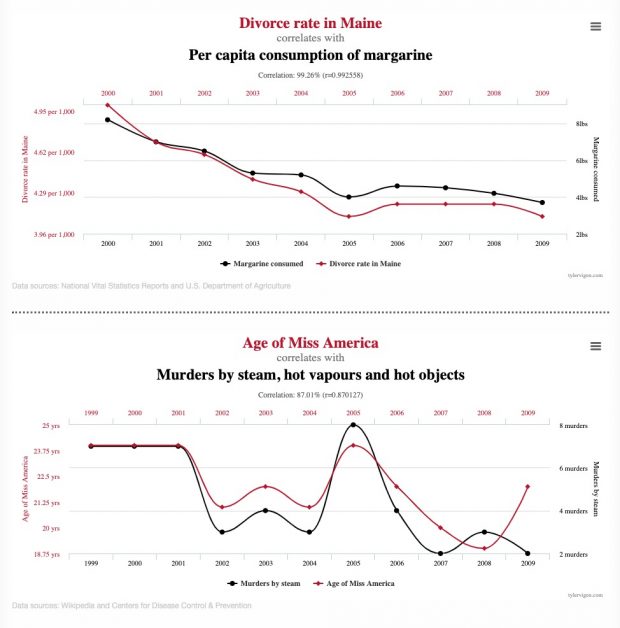
Wozu es führt: lauter sinnlose Korrelationen
https://www.technologyreview.com
Hundreds of AI tools have been built to catch covid. None of them helped.