Although the authors of this new JACI paper claim to having done a systematic review they even missed two key papers that even contradict the statement above Continue reading Will they ever read the literature?
Although the authors of this new JACI paper claim to having done a systematic review they even missed two key papers that even contradict the statement above Continue reading Will they ever read the literature?
Using data described in Science Magazine (basically 4% of all published books) I constructed a plot showing an update to the 100 year old mystery why we get allergic to certain substances.
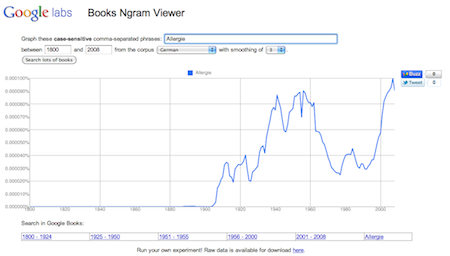
There seems to be a good chance for 2011 if the line will further increase…
There is a new Edge Special Event about the Hillis’s question “WHO GETS TO KEEP SECRETS?”
The question of secrecy in the information age is clearly a deep social (and mathematical) problem, and well worth paying attention to.
When does my right to privacy trump your need for security?; Should a democratic government be allowed to practice secret diplomacy? Would we rather live in a world with guaranteed privacy or a world in which there are no secrets? If the answer is somewhere in between, how do we draw the line?
With all the wikileaks hype over the last year, the Edge essay is la perfect supplement to our last paper about anonymity in genetics – check out BMC Ethics “Caught you: Threats to confidentiality due to the public release of large-scale genetic data sets“.
What we didn’t mention in this paper are more complicated statistics like stochastic record linkage – more on that in RJournal 2/2010, p.61 ff
It seems that I am not alone here to think that the hygiene hypothesis has thrown back allergy research for 20 years – despite the desperate attempts of journalists and scientists. Here is an an excerpt of Hygiene hypothesis: wanted—dead or alive
When it comes down to all, the best evidence to prove or dispute the hygiene hypothesis will probably come from ongoing and future randomized trials of interventions, e.g. treatment with probiotics and microbial products, that have been developed in the light of the hygiene hypothesis. In the mean time, we must prepare ourselves to face the results of these trials as well as of other types of evidence. It is a possibility that it may turn out that the hygiene hypothesis is more dead than alive, or at the least needs another revision.
We just published the largest study so far of human serum cytokines providing for the first time reference values.
In this study we investigated serum samples from 944 individuals of 218 asthma-affected families by a multiplex, microsphere based system detecting at high sensitivity eleven asthma associated mediators: eotaxin (CCL11), granulocyte macrophage stimulating factor (GM-CSF), interferon gamma (IFNγ), interleukin-4 (IL-4), IL-5, IL-8, IL-10, IL-12 (p40), IL-13, IL-17 and tumor necrosis factor alpha (TNFα). Continue reading The largest study so far on serum cytokines
There is a new database that describes itself as
a database of extremely similar Medline citations. Many, but not all, of which contain instances of duplicate publication and potential plagiarism. Deja vu is a dynamic resource for the community, with manual curation ongoing continuously, and we welcome input and comments.
Continue reading Déjà vue database finally
It stirred already the vitamin D community, the new dietary reference intakes with comments at newspapers and blogs. But read yourself all 999 pages ;-) Continue reading Finally: more qualified dietary reference intakes for vitamin D
This is not a joke – we can easily drop the last 3 or 4 nature genetics volumes for ignorance of basic facts. I have written here many times about the usefulness of current GWAs but missed the details of a Cell paper by McClellan & King that I am wholeheartedly supporting (although not all other science bloggers) Continue reading Drop the last 3 nature genetics volumes from the libraries
I haven’t seen the ceremony award 2010 so far but read of a prize for discovering that symptoms of asthma can be treated with a roller-coaster ride (Simon Rietveld of the University of Amsterdam, The Netherlands, and Ilja van Beest of Tilburg University, The Netherlands) and of the management prize for demonstrating mathematically that organizations would become more efficient if they promoted people at random (Alessandro Pluchino, Andrea Rapisarda, and Cesare Garofalo of the University of Catania, Italy), yea, yea.
There is a new NEJM paper on asthma genetics.
I am not really sure if I am really an author of this paper ( I am listed as collaborator whatever that means). At least, the study relies heavily on thousands of samples and a big data set that I sent to the study management board) while it its certainly a paper that summarizes the efforts of many years of research.
Unfortunately it shares all problems of current GWAs studies. As I did not have any chance to comment on it during the preparation, here are some thoughts (and it is funny to see that my blog is ranked higher in Google search than the original paper- a curiousity showing a changing world where scientists have already lost their power on public opinion). But lets work along the abstract.
Susceptibility to asthma is influenced by genes and environment; implicated genes may indicate pathways for therapeutic intervention.
The first sentence is a bit trivial. I usually delete such sentences from manuscripts. The second sentence is a mantra repeated over and over again, never proven, origininating from application prose than from any scientific study.
We carried out a genomewide association study by genotyping 10,365 persons with physician-diagnosed asthma and 16,110 unaffected persons, all of whom were matched for ancestry. We used random-effects pooled analysis to test for association in the overall study population and in subgroups of subjects with childhood-onset asthma (defined as asthma developing before 16 years of age), later-onset asthma, severe asthma, and occupational asthma.
I wonder why we need now >25,000 DNAs as Coookson already reported in his book that “he found the asthma gene” (ref)? Are effects are so small (or is the design of genotyping chips so poor) that we need so many samples? The introduction says that current GWAS technology is the method of choice for identifying genes that influence complex disease.
I do not agree – we need better defined phenotypes than just some self reported disease (separating all kind of “asthmatic” obstruction). We need SNP alleles of the whole frequency range (not just the tagging SNPs), simply speaking better and not just larger studies.
We observed associations of genomewide significance between asthma and the following single-nucleotide polymorphisms: rs3771166 on chromosome 2, implicating IL1RL1/IL18R1 (P=3×10(?9)); rs9273349 on chromosome 6, implicating HLA-DQ (P=7×10(?14)); rs1342326 on chromosome 9, flanking IL33 (P=9×10(?10)); rs744910 on chromosome 15 in SMAD3 (P=4×10(?9)); and rs2284033 on chromosome 22 in IL2RB (P=1.1×10(?8)). Association with the ORMDL3/GSDMB locus on chromosome 17q21 was specific to childhood-onset disease (rs2305480, P=6×10(?23)). Only HLA-DR showed a significant genomewide association with the total serum IgE concentration, and loci strongly associated with IgE levels were not associated with asthma.
Although not referenced in the paper, we described an association with the IL1 cluster already in 1994 that I further refined in 2008 to IL18R1 which was also published in our NG paper 2009.
IL13, IL33, HLA-DQ, IL2RB: nothing new under the sun. Also the “ORMLD3” story has been fully published (see some earlier comments), while ORMLD3 doesn’t even appear in table 2 (reported are only results for GSDMA and GSDMB). For any unknown reason ORMDL3 makes it into the last sentence of the paper and the abstract. I have no idea how in such highly correlated number space, a forward stepwise regression can establish an independent association. Ordering results by p value, does that make any sense?
What worries me: There was non interaction between SNP variants, which would indicate that a single gene alone could lead to asthma – something extremely unlikely from all previous studies where we could never find a monogenic asthma form.
CONCLUSIONS: Asthma is genetically heterogeneous. A few common alleles are associated with disease risk at all ages. Implicated genes suggest a role for communication of epithelial damage to the adaptive immune system and activation of airway inflammation. Variants at the ORMDL3/GSDMB locus are associated only with childhood-onset disease. Elevation of total serum IgE levels has a minor role in the development of asthma.
Where is the longstanding claim of Cookson of IgE and FCER1B? Gone? The current NEJM article even refutes the January NEJM issue of DENND1B (largely by the same authors: “none of these associations were significant after correction for multiple comparisons”). What will be the content of the next paper in 2020 with 100K asthmatics?
The little overlap between “IgE loci” and “asthma loci” is not an argument that “elevation of IgE level is an inconstant secondary effect of asthma” as also this new paper is just a cross-sectional study and these are just some association, nothing else.
It drives me crazy – the new JACI review with a detailed explanation how VDD (vitamin D deficiency) induces food allergy (FA). Unfortunately, it has never been show that FDD is related to FA – while the opposite may be true… Continue reading What the #$*! Do We Know!?
I wasn’t sure whether to continue here the 2008 dung hill blog post or just opening a new thread as a colleague just entered my office with some news. Continue reading More dung hills
Here comes the most striking connection between calcium and IgE that updates some earlier posts here
The fact that the regulation of intracellular Ca2+ concentration is different in various T-cell effectors may offer the opportunity to target key intermediates … to inhibit specifically the functions of one given T-cell subset. Continue reading Once more: Calcium and IgE
I need to refer here to a post 3 years ago and the medical literature that genes frequencies may have changed rapidly between generations.
Any empirical proof of this hypothesis, however, is scarce so far. Or I have to say, until this week, when I found a study published earlier in PLoS ONE that tackles this problem: Selection for Genetic Variation Inducing Pro-Inflammatory Responses under Adverse Environmental Conditions in a Ghanaian Population Continue reading Genes on the fast lane
Two recent studies used food frequency questionnaires to predict vitamin D status and later allergy (Devereux 2007 and Camargo 2007) probably the only two studies that seem to contradict the vitamin D hypothesis.
New research now reported at the ATS congress [Poster Board # A84] “Measurement of Vitamin D Levels Utilizing Laboratory and Dietary Recall Information from the Tennessee Children’s Respiratory Initiative” and published in Am J Respir Crit Care Med 181;2010:A1890 shows that FFQs don’t predict vitamin D status Continue reading Why FFQs don’t predict vitamin D status