More than 7 years ago, I wrote a blog post that there is nothing new under the sun predicting the next asthma genetics study for 2020 to include 100K asthmatics.
Ok, I am wrong the paper appeared already a week ago with 360K asthmatics while basically doubling the number to 136 independent risk variants. It’s not an asthma only study as the authors had a rather loose definition of asthma or rhinitis or eczema – is is more about the allergy/atopy complex.
It will be a long time reading and replicating the data while my first interest was to examine the affected gene list as shown in table ST15. Using the online Enrichr database I get the following result from the GO ontology
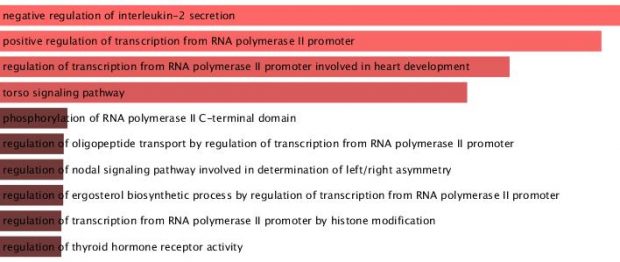
The highest combined score is with a negative regulation of IL-2. What does that mean? A genetically disturbed pathway predicted by a gene ontology network?
IL2 was discovered in 1976 by Robert Gallo by growing T-cells in culture for more than nine months by stimulating lymphocytes with phytohemagglutinin. ((IL1 was described back in 1972 by Charles Dinarello). Gallo identified T-cell growth factor (TCFG), now known as interleukin-2 (IL-2) as being absolutely important for a protective immune responses (Science 1976).
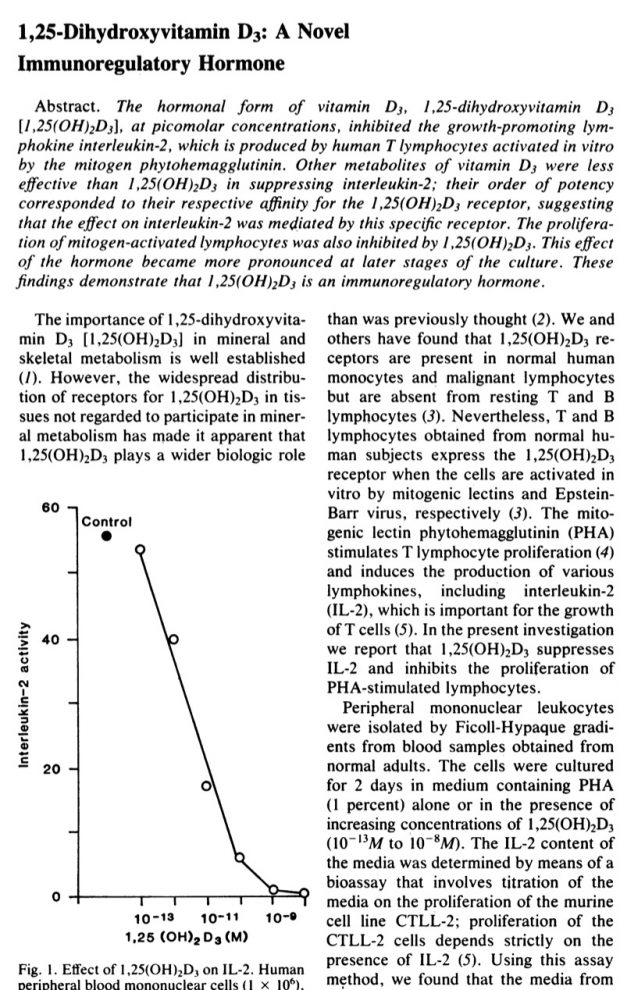
The next important paper was also published in Science 1984 showing the following figure
So we have in allergy 2 hits on IL2: by genetics (as show in the new study) and by early vitamin D supplementation (as I reviewed earlier). Maybe we need more clinical studies like the Zhang 2016 study:
In a general population-derived birth cohort, we found that in infants who developed food allergy, cord blood displayed a higher monocyte to CD4(+) T cell ratio and a lower proportion of natural regulatory T cell (nT(reg)) in relation to duration of labor. CD14(+) monocytes of food-allergic infants secreted higher amounts of inflammatory cytokines (IL-1β, IL-6, and tumor necrosis factor-α) in response to lipopolysaccharide. In the presence of the mucosal cytokine transforming growth factor-β, these inflammatory cytokines suppressed IL-2 expression by CD4(+) T cells. In the absence of IL-2, inflammatory cytokines decreased the number of activated nT(reg) and diverted the differentiation of both nT(reg) and naïve CD4(+) T cells toward an IL-4-expressing nonclassical TH2 phenotype.
Suppressed IL2 is a key for allergy development shown also in experiments by Bonnet 2016:
We previously demonstrated that Tregs can be selectively expanded and activated by low doses of IL-2 (ld-IL-2) inducing immunoregulation without immunosuppression and established its protective effect in autoimmune diseases. In this study, we evaluated the ability of ld-IL-2 to control allergy in an experimental model of food allergy. Ld-IL-2 induced Treg expansion and activation that elicited protection against clinical manifestations of food allergy in two mouse models with OVA and peanut. This clinical effect was lost in Treg-depleted mice, demonstrating the major contribution of Tregs in ld-IL-2 efficacy. Mechanistic studies further indicated that protection from allergy could be explained by a Treg-dependent local modification of the Th1/Th2 balance and an inhibition of mast cell recruitment and activation. Preventive and therapeutic effects of ld-IL-2 were observed over a 7-mo-period, highlighting its long-term efficacy. This study demonstrated that ld-IL-2 is efficient to prevent and to treat allergic immune responses, and thus represents a promising therapeutic strategy for managing allergic diseases.
What we do need now for allergy prevention is a trials of newborns with low dose IL2 and also more trials refining the IL2 antagonist application of vitamin D.