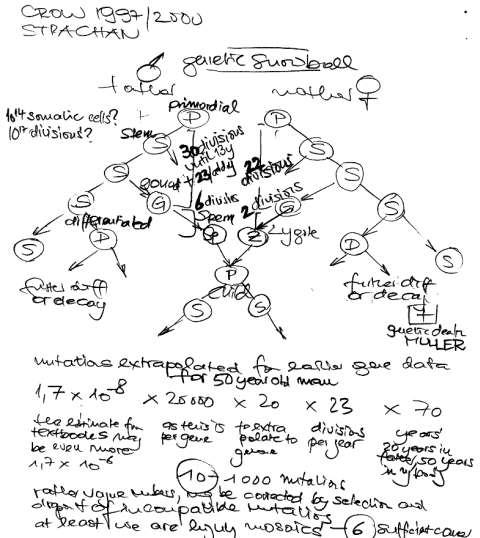
We have a new paper at Sci Rep online “High degree of polyclonality hinders somatic mutation calling in lung brush samples of COPD cases and controls“.
It took a long time from my initial grant application at Sander Stiftung in Dec 2009 (where it was rejected), to the field work within the scope of the EvA study (where the PI Loems Ziegler-Heitbrock retired).
Followed by some first analysis together with Francesc at CNAG in Barcelona the final publication now appeared – my gratulations to Gian-Andri and Ivo Gut for their hard work!
Chronic obstructive pulmonary disease (COPD) is induced by cigarette smoking and characterized by inflammation of airway tissue. Since smokers with COPD have a higher risk of developing lung cancer than those without, we hypothesized that they carry more mutations in affected tissue.
We called somatic mutations in airway brush samples from medium-coverage whole genome sequencing data from healthy never and ex-smokers (n=8), as well as from ex-smokers with variable degrees of COPD (n=4). Owing to the limited concordance of resulting calls between the applied tools we built a consensus, a strategy that was validated with high accuracy for cancer data.
However, consensus calls showed little promise of representing true positives due to low mappability of corresponding sequence reads and high overlap with positions harbouring known genetic polymorphisms. A targeted re-sequencing approach suggested that only few mutations would survive stringent verification testing and that our data did not allow the inference of any difference in the mutational load of bronchial brush samples between former smoking COPD cases and controls.
So we would have probably needed a higher genome coverage on our brush sample mix. Or should we have sequenced more single cells as discussed in the paper?
At least, we now know, that sequencing at rather low coverage rate is not a screening tool for expected cancer development. Are there less pre-malignant lesions than expected? When looking at some other papers (Cancer Genome Atlas, esophagus, and more recently colon samples, I can only confirm what Iñigo Martincorena wrote
this study emphasizes how little we know about somatic evolution within normal tissues, a fundamental process that is likely to take place to varying degrees in every tissue of every species.
Somatic mutations accumulates with age. There may be even more mutations in the aging esophagus than in sun-exposed human skin. Lee-Six estimates 43.6 mutations /year, while I still have a gut feeling that there is no gradually accumulation of mutations (until the second hit) but a clonal expansion of a single
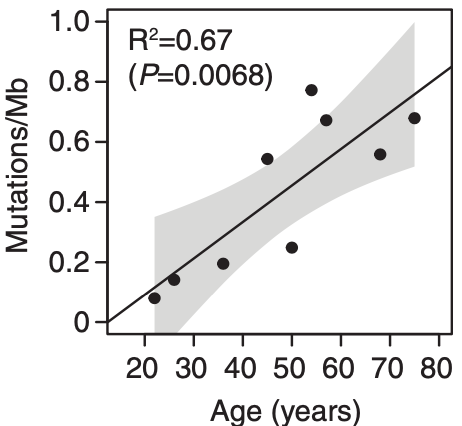
bronchial cell, hit by a single smoke stream. With this hypothesis, smoking would not kill by accumulation of deleterious mutations, but by the never ending re-exposure until the ultimate deleterious mutation occurs.
Many more of these timeline studies will be necessary to explain why the lung cancer risk drops immediately after you stop smoking.