or in more scientific terms: Why are F1 hybrids so often sterile or lethal? The Dobzhansky-Muller theory says that there is an incompatibility between genes with reduced fitness that have diverged between species. So far nobody has ever observed a D-M gene but a new Science paper describes two genes that separate D. simulans and D. melanogaster: lhr (lethal hybrid rescue) and hmr (hybrid male rescue). Yea, Yea.
Category Archives: Genetics
For the first time two human genomes compared
Another “first discovery” in this nature genetics preprint although the analysis could have already been done some years earlier. The CNV specialists from Toronto now compare the Human Genome Project sequence with the Celera sequence – the gap between the two compilations was obviously bigger than the intra-sequence gaps. Of course both sequences are still mosaics from several individuals but the analysis nicely exemplifies how difficult it will be to compare the genome of two different human beings.
The authors employ a whole battery of alignment tools BLAT, MEGABLAST, GCA and A2Amapper. Of course results depend on the strategy, definition and implementation. As show by FISH analysis most of the discrepancies are true and can be classified into a few categories – insertions or deletions if seen from the second genome (has somebody ever thought about a minimal human genome?), mismatches and inversions. We are getting here a preview of the diagnostic workup in a patient in 2026. This blog contains forward looking statements while the responsibility rests solely with the reader. Yea, yea.
Headline: For the first time 2 human genomes compared
What sounds brand new in a current nature genetics preprint, could have even been done already a couple of years before. The CNV specialists from Toronto compare now the Human Genome Project sequence with the Celera sequence. Was the gap between these two compilations bigger than the intra sequence holes? Both sequences are of course mosaics from several individuals but the analysis nevertheless exemplifies how difficult it may be to compare even the genome of two related individuals. The authors employ a whole battery of alignment tools BLAT, MEGABLAST, A2Axxx, GCA. Of course results are different depending on strategy, definition and implementation used. As show nicely by FISH analysis most of the discrepancies are true and can be classified into a few categories – insertions with and without corresponding fragment or deletions if seen from the second genome (has somebody ever thought about a minimal human genome?), mismatches and inversions. We are getting here a preview of the diagnostic workup in a patient in 2026. This blog contains forward looking statements while the responsibility rests solely with the reader. Yea, yea.
New LD measure
There is a new way to calculate LD that may overcome the limitations of D’ and R^2 that are not easily generalizable to multiallelic markers (or haplotypes) and depend on the distribution of SNPs (or haplotypes).
The paper is at BMC, the sources at the authors’ website. I have slightly modified the program to allow input and output file names on the command line before compiling it. Use at your own risk, yea, yea.
New R packages for SNP studies
Tit-for-tat or altruism in science
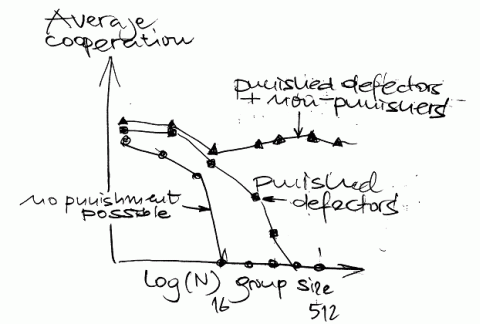
No, this essay will not deal with altruism in science but with the science of altruism. There are two new papers from the Fehr group (one in Science on Nov, 3 about diminished reciprocal fairness after magnetic stimulation of the right prefrontal cortex and a second in Nature on Aug, 24 about altruism in two indigenous groups in Papua New Guinea). I was, however, much more impressed by their recent review of human altruism.
Cooperation between genetically unrelated groups is a typical human behaviour (otherwise seen only in ants, bees and the naked mole rat) where there seems a strong reciprocity between selfishness and altruism. Cooperation is rarely stable and may deteriorate under worse conditions. Altruistic rewarding and reputation seeking seem to be the most powerful determinants of future donors’ behaviour where effects of punishing behaviour seem to be underestimated: Cooperation in larger group continues only if punishment of defectors and non-punishers is possible.
An utter refutation
I am slow in commenting on a paper that has already been published earlier this year – Joe Terwillingers vivid refutation of the fundamental theorem of the hapmap proponents that
if a marker is in tight LD with a polymorphism that directly impacts disease risk, as measured by the metric r^2, then one would be able to detect an association between the marker and disease with sample size that was increased by a factor of 1/r^2 over that needed to detect the effect of the functional variant directly
I cannot comment on the statistical proof but fear from my recent experience with Crohn and asthma tags that he may be right with his assumption: Even marker in high LD with the functional variant may not show any association at all. These may be bad news for all those currently running large screening programs with hapmap based variants believing that P(A|BC)=P(A|Bc)=P(A|B), yea, yea.
Addendum
Tag SNPs also do not work with CNVs
Who will survive?
When looking at gene variants in a population we may forget that even having a perfect sampling scheme this will not be an unbiased view of the human genome. Earlier studies suggested that up to 75% of conceptions are lost during early development; a further indicator of an biased view are unexplained cases of departure from Hardy-Weinberg equilibrium.
Selective survival during early pregnancy is still a terra incognita and except of studies in the Hutterites I am not aware of any (modern) study that looked at selective survival.
A study of Grant Montgomery now shows fresh data on genomewide allele sharing in 1,592 DZ twins from Australia and 336 DZ pairs from the Netherlands.
It is somewhat disappointing that there is no excess allele sharing in the HLA region nor somewhere else in the genome. Maybe further studies can do that a high resolution than with just 359 microsatellite marker?
Escaping from a swamp
The November AJHG has an excellent re-analysis of the dysbindin-schizophrenia association using new methodology that surpasses all previous meta-analysis techniques. As the single SNP association results from the previous 6 studies cannot be directly compared, they construct a European super-hap map from all tag SNPs in that region, place them in a phylogenetic tree before finally mapping all single associations on these haplotypes. Their Fig.1B show the main results; as the circles in Fig.1B are somewhat confusing, I have withdrawn their results – adding the haplotype frequencies and ordering the studies by year of publication.
We may think of a triple-blind study – neither patients, nor PIs, nor we did know anything before. The results are alarming. I do not understand how the Kirov set could have included all haplotypes and why the Schwab/Williams set is in opposition to the Straub/Bogaert/Funke set.
What could have gone wrong? The authors of the current re-analysis believe that population differences are an unlikely reason for the inconsistency as the allele frequencies match between studies. Good news that genotyping errors may be largely excluded.
Unfortunately the authors remain vague why there is no common causal variant. Have there been different sampling schemes, different diagnostic thresholds, different environmental exposures in the previous studies? Is dysbindin at all a schizophrenia gene, or only under a certain genetic background? It seems possible that studies of one branch are false positives. Or is the haplotype reconstruction in the re-analysis erroneous for whatever reasons?
Von Münchhausen is well know for escaping from a swamp by pulling himself up by his own hair. I would like I could do that too.
Number cruncher
In a recent blog I described high resolution SNP datasets that are available on the net. To work with these datasets you will probably need to upgrade your hardware and software. For data handling many people stick nowadays to commercial SQL databases that have plugins for PD software.
My recommendation is to save that money and store the data in a special format that may be more useful for these large dataset; details are in a technical report that I will upload later this day. In the meantime you can already check some software tools to work with these large datasets. This is what I know so far
- David Duffy has recompiled his sibpair program |link
- Geron(R) has something under development |link
- Jochen Hampe and colleagues offer Genomizer |link
- Franz Rüschendorf developed Alohomora |link
- I renember about SNPGWA, a development at Wake Forest University |no link yet
- there will be a R-Bioconductor package by Rob Scharpf |no link yet
- R library GenABEL by Yurii Aulchenko |link
- R library SNPassoc by Juan González |link
Addendum
A technical report how to work with large SNP dataset is now also available at my paper section. Alternatives to what I am suggesting in this paper, have been set out by an anonmyous reviewer
For R users, if SQLite limits are reached, hdf5 (http://hdf.ncsa.uiuc.edu/HDF5/) may be one way forward for really huge table structures since there is an R interface already available. PostgreSQL column limit depends on data type with a maximum of 1600 for simple types. MySQL with the BerkeleyDB backend may be like SQLite with no obvious column count limit. Metakit is not mentioned – it is column oriented and probably also has “unlimited†columns as long as each database is < 1GB or so.
What people search for
“Dissecting the complex genetic basis of mate choice” is the lengthy title of a lengthy text that tells us
males produce complex signals and displays that can consist of a combination of acoustic, visual, chemical and behavioural phenotypes…
The authors come from a school of integrative biology. I wonder why they have missed the excellent work in humans on HLA, fertility and mate choice.
Having said that, I would even suggest a radical different approach by looking at “What people search for” – hopefully I get now also hits on my blog for Paris Hilton, Renee Zellweger, Britney Spears, Heidi Klum, Pamela Anderson, Jessica Simpson and Jennifer Lopez ;-) Dissecting the complex genetic basis of mate choice shouldn´t be as complicated as you may imagine from this nature reviews genetics paper, yea, yea.
Better than the Delphi oracle
A new paper shows a nice workflow how to do an in vitro prediction which drug will suppress a certain tumor. The authors are simply linking the phenotype of the cell line “50% inhibitory concentration by drug X” with its expression signature. The good news are that doing both in one vial (phenotyping and expression analysis) is leading to excellent results.
Is there any trick to do this also system-wide e.g. for the metabolism of a substance and its signalling pathway? Pharmacogenetics would greatly benefit from such an approach, nay, nay.
In the heat of the night
Gene lists by automatic literature extraction
Just found at the HUM MOLGEN bulletin board a link to Fable, a new automated literature extraction system. Fable is pretty fast and can output gene lists. Sure, the screenshot below shows only those genes that I mentioned in the abstract, but this is not so bad as the most important genes wil be placed there.
BTW, the number of reviews on asthma genetics have been falling to less than 50% after closing the Asthma Gene Database. Maybe this new service will help to re-establish the former output of reviews ;-) yea, yea.
IL4 cluster revisited
I am interested in 5q31 and the IL4 cluster since I met David Marsh in the lobby of a hotel in Heidelberg around 1993. David was one of the founding fathers of asthma genetics and I renember how he vividly told me that he has a forthcoming Science paper on the IL4 cluster and IgE. The cluster is still one of the best allergy regions where the signalling through IL4 and IL13 now gets more interest than the work of any of his competitors.
Nature genetics now has an update on the 3-dimensional resolution of the genomic region. It is not cristallographic work as might be expected but a nice study of the chromatin structure that is leading to a coordinated expression of these cytokines. SATB1 (special AT-rich sequence binding protein 1) is thought to anchor specialized sequences letting DNA loops come into interaction. I wonder if there might be even a direct physical interaction of the IL4 and IL13 promotor and if there will be any SNP influencing that interaction? David (who died of brain cancer in 1998) would have really liked this work. Yea, yea.