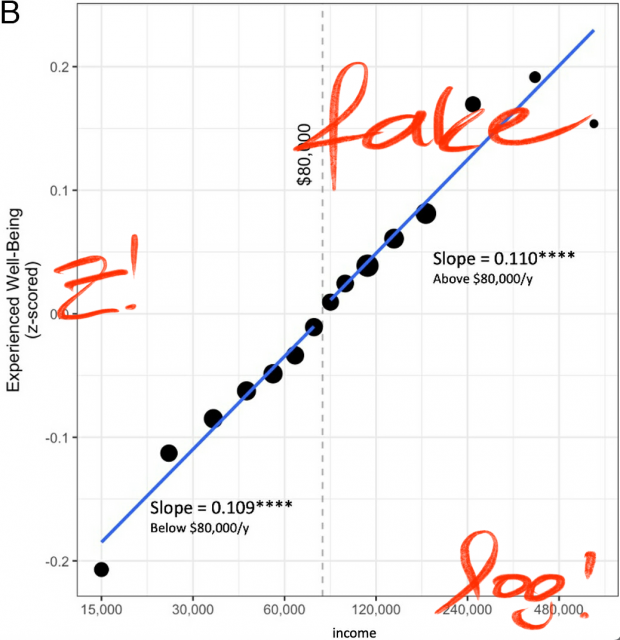
ephorie.de has a convincing re-analysis that Daniel Kahneman is wrong, who purportedly demonstrated that emotional wellbeing increases with income but plateaus at $75,000…
 .
.
ephorie.de has a convincing re-analysis that Daniel Kahneman is wrong, who purportedly demonstrated that emotional wellbeing increases with income but plateaus at $75,000…
 .
.
Jim Baggott quotes John Heilbron, a Kuhn scholar, on the question what is a scientific myth
A scientific myth is not produced by accident or error. It requires effort. “To qualify as a myth, a false claim should be persistent and widespread,” Heilbron said in a 2014 conference talk. “It should have a plausible and assignable reason for its endurance, and immediate cultural relevance,” he noted. “Although erroneous or fabulous, such myths are not entirely wrong, and their exaggerations bring out aspects of a situation, relationship or project that might otherwise be ignored.”
Ich habe es dem OB von München 1993 in einem Brief vorgeschlagen, aber er hat mir geantwortet, daß es politisch nicht durchsetzbar sei. 31 Jahre später ist es nun so weit…

Ob es eine gute Idee war? Die Studienlage ist ja immer noch nicht gut da bisher nur örtlich und zeitlich begrenzte Tests durchgeführt wurden.
Denn natürlich gibt es auch Sekundäreffekte etwa dass die insgesamt gefahrene Strecke sinkt wenn für viele Autopendler die Attraktivität sinkt. Warum nicht gleich auch noch die Entfernungspauschale streichen?
Mikael Laakso, Lisa Matthias, Najko Jahn
Open is not forever: a study of vanished open access journals.
JASIST 2021;72:1099-1112
The preservation of the scholarly record has been a point of concern since the beginning of knowledge production. … We found 174 OA journals that, through lack of comprehensive and open archives, vanished from the web between 2000 and 2019
Zenodo has the full dataset
| Journal Name | Publisher | Discipline |
|---|---|---|
| International Journal of Information Technology | World Academy of Science, Engineering and Technology | Computer Science |
| Journal of Mundane Behavior | Millersville University | Sociology |
| Complexity International | Johnston Center, Charles Sturt University | Complex Systems |
| Pharmaceutical Reviews | Pharmainfo.net | Pharmacology |
| Annales Universitatis Mariae Curie-Sklodowska - Sectio D. Medicina | Uniwersytet Marii Curie-Sklodowskiej | Medicine |
| Essays in Education | Columbia College * Department in Education | Education |
| Peace Tourism Journal | International Centre for Peace Through Tourism Research | POLITICAL SCIENCE - INTERNATIONAL RELATIONS, TRAVEL AND TOURISM |
| Journal of Global Social Work Practice | Dominican University * Graduate School of Social Work | SOCIAL SERVICES AND WELFARE |
| International Quarterly of Sport Science | Hungarian Society of Sport Science | SPORTS AND GAMES |
| Public Knowledge Journal | Public Knowledge Journal | Political science |
| Hamburg Review of Social Sciences | Hamburg Review of Social Sciences | Social Sciences |
| Doping Journal | Doping Journal | BIOLOGY - BIOCHEMISTRY, PHARMACY AND PHARMACOLOGY |
| eMinds | Universidad de Oviedo * Departamento de Informatica | COMMUNICATIONS, COMPUTERS |
| Crossroads | Alta Scuola di Economia e Relazioni Internazionali | BUSINESS AND ECONOMICS, LAW, POLITICAL SCIENCE - INTERNATIONAL RELATIONS |
| Electronic Journal of Organizational Virtualness | Journal of Organizational Virtualness | COMPUTERS - INTERNET |
| AntePodium | Victoria University of Wellington * School of Political Science and International Relations | POLITICAL SCIENCE |
| Durham Anthropological Journal | University of Durham * Department of Anthropology | ANTHROPOLOGY |
| Studencheskii Nauchnyi Zhurnal | Studencheskii Nauchnyi Zhurnal | SCIENCES: COMPREHENSIVE WORKS |
| Learning Exchange | University of Westminster | EDUCATION - HIGHER EDUCATION |
| Romanian Medical Reviews & Research | Viata Medicala | MEDICAL SCIENCES |
| Kuwait Radiology Journal | MedicalMantra Information Systems | MEDICAL SCIENCES - RADIOLOGY AND NUCLEAR MEDICINE |
| Transformations (Atlanta) | Associated Colleges of the South | HUMANITIES: COMPREHENSIVE WORKS - COMPUTER APPLICATIONS |
| Afroeuropa | Universidad de Leon * Departamento de Filologia Moderna | Linguistics |
| IBScientific Journal of Science | IBScientific Publishing Group | Engineering |
| Anatolian Journal of Obstetrics & Gynecology | Alkim Basin Yayin Ltd. ti. | Gynecology and Obstetrics |
| Journal of Education for International Development | Educational Quality Improvement Program | Education |
| Rocky Mountain Communication Review | University of Utah * Department of Communication | COMMUNICATIONS |
| Biomirror | Xinnovem Publishing Group | Biology |
| Journal of Advanced Pharmaceutical Research | Pharmaceutical Research Foundation | Pharmacology |
| Working Papers in Art and Design (Online) | University of Hartfordshire * Department of Design and Foundation Studies | Art |
| Neurobiology of Lipids | Neurobiology of Lipids | BIOLOGY - PHYSIOLOGY, MEDICAL SCIENCES |
| Journal of Dagaare Studies | University of Hong Kong | HISTORY - HISTORY OF AFRICA, LINGUISTICS, LITERATURE |
| International Journal of Pharmacy Education | Samford University * McWhorter School of Pharmacy | EDUCATION - HIGHER EDUCATION, PHARMACY AND PHARMACOLOGY |
| International Journal of Pharmaceutical and Biomedical Research (IJPBR) | PharmSciDirect Publications | Pharmacology |
| School of Advanced Technologies. Electronic Journal | Asian Institute of Technology | Computers |
| Lyon Pharmaceutique (Online) | Association des Pharmaciens - Anciens Eleves - Amis de la Faculte de Pharmacie de Lyon | Medicine |
| St. Francis Journal of Medicine | St. Francis Medical Center | Medicine |
| Ciência & Tecnologia dos Materiais | Sociedade Portuguesa de Materiais | General and Civil Engineering |
| Social Science Paper Publisher | University of Western Ontario, Department of Sociology | Social Sciences |
| The Carleton University Student Journal of Philosophy | Carleton University, Department of Philosophy | Philosophy |
| African Journal of Environmental Assessment and Management | AJEAM/RAGÉE | Environmental Sciences |
| Cal Poly Journal of Interdisciplinary Studies | California State Polytechnic University | Education |
| Philosophic Nature | Excogitation & Innovation Laboratory | Science (General) |
| Asia-Pacific Journal of Oncology & Hematology | San Lucas Medical | Medicine |
| International Journal of Signal Processing | World Academy of Science, Engineering and Technology | Electrical and Nuclear Engineering |
| Advances in mathematical and computational methods | Information Engineering Research Institute | Mathematics |
| Social Science Letters (SSL) | Information Engineering Research Institute | Social Science |
| International journal of swimming kinetics | International Society of Swimming Coaching | Sports physiology, sports medicine |
| Open source science journal | Academy of Economic Sciences | Computer Science |
| Revista sujeto, subjetividad y cultura | School of psychology University of Arts and Social Sciences, ARCIS, Santiago de Chile | Psychology |
| Biotechnology and health sciences | Qazvin Qazvin University of Medical Sciences | Biotechnology and Health Sciences |
| European Journal of Economic & Political Studies | Faith University | |
| Technológia Vdelavania | Združenie Slovdidac | Education |
| Mukaddimah: jurnal studi Islam dan informasi PTAIS | Kopertais Wilayah III | Religion |
| New theology review | Liturgical Press | Theology |
| Journal of Medical Sciences Research | Journal of Medical Sciences Research (J M S R) | Medicine |
| Revista de Educao e de Tecnologia Aplicadas Aeronutica | Comando da Aeronutica | Transportation |
| Patria Grande : Revista Centroamericana de Educacin | Coordinacin Educativa y Cultural Centroamericana | Education |
| Ovid, Myth and (Literary) Exile | Ovidius University Press | Languages and Literatures |
| Journal of eLiteracy | University of Glasgow | Library and Information Science, e-literacy |
| Informacini Technologij Taikymas vietimo Sistemoje | Kauno Kolegija (Kaunas University of Applied Sciences) | Education |
| Perspectives in Oral Sciences | Universidade Positivo | Dentistry |
| Acta Monographica | Acta Monographica | SCIENCES: COMPREHENSIVE WORKS |
| Journal of Advances in Developmental Research | Gujarat Vidyapeeth University | Sociology |
| Interdipendenze : Rivista di Teoria e Ricerca Sociale, Studi Ecologici, Etnoscienze | Societ di Etnosociologia e Ricerca Sociale (S.E.Ri.S) | Environmental Sciences --- Sociology |
| Lianes | Lianes Association | Sociology |
| E-journal of Reservoir Engineering | Petroleum Journals Online | Mining and Metallurgy |
| Social Justice in Context | Institute for Social Justice, East Carolina University | Social Sciences |
| Educao Profissional : Cincia e Tecnologia | Servio Nacional de Aprendizagem Comercial do Distrito Federal | Science (General) --- Education |
| SRX Biology | Scholarly Research Exchange | Biology |
| More than Thought : a Scholarly Literary Journal Devoted to Consciousness | More than Thought | Languages and Literatures |
| European Journal of Clinical & Medical Oncology | San Lucas Medical | Oncology |
| European Neurological Journal | San Lucas Medical | Neurology |
| Journal of Neuroparasitology | Ashdin Publishing | Internal medicine --- Neurology |
| Scholars' Research Journal | Wolters Kluwer | Biotechnology |
| Arquivstica.net | Arquivstica.net | Library and Information Science |
| Emergent Australasian Philosophers | Dean Goorden and Matthew Paul, Ed. & Pub. | Philosophy |
| Journal of Coagulation Disorders | San Lucas Medical | Internal medicine |
| Comunicaciones de la Sociedad Malacolgica del Uruguay | Sociedad Malacolgica del Uruguay | Zoology |
| Ciencias Sociales Online | Universidad de Vina del Mar * Escuela de Ciencias Sociales | Social Sciences |
| eJournal of Biological Sciences | EJARR Publishing | Biology |
| Global Tourism | Global Tourism Consulting and Training | Geography --- Social Sciences |
| Translocations : Irish Migration, Race and Social Transformation Review | Dublin City University | Migration --- History |
| Cancerologa : Revista del Instituto Nacional de Cancerologa | Instituto Nacional de Cancerologa de Mxico | Oncology |
| Analytica | Analytica | Philosophy |
| Open Aging Journal | Bentham open | Internal medicine |
| Research in Biotechnology | School of Bio Sciences and Technology VIT University | Biotechnology |
| War Crimes, Genocide and Crimes Against Humanity | Penn State Altoona | Social and Public Welfare --- Law |
| Research Journal of International Studies | European Journals, Inc. | Social Sciences --- Political Science |
| Journal of Preventive Medicine | Institute of Public Health, Iasi | Public Health |
| Middle East Studies | Middle East Studies | Social Sciences |
| Journal of Forensic Accounting | R.T. Edwards, Inc. | Accounting |
| Gendernye Issledovani | Kharkov Center for Gender Studies | Gender Studies |
| Electronic Journal of Environmental, Agricultural and Food Chemistry | Universidade de Vigo | Nutrition and Food Sciences --- Agriculture (General) |
| Electronic Journal of Literacy through Science | University of California, Davis | Education |
| Tierra Tropical : Sostenibilidad, Ambiente y Sociedad | Universidad EARTH | Environmental Sciences |
| Information Technology, Learning, and Performance Journal | Organizational Systems Research Association | Computer Science --- Education |
| International Diabetes Monitor | Medical Forum International | Internal medicine |
| Journal of Azerbaijani Studies | Khazar University | Social Sciences |
| Nordic Notes | Flinders University Adelaide | Ethnology --- History |
| Journal of dentistry, oral medicine and dental education | Scientific journals international | Dentistry |
| Journal of Research Methods and Methodological Issues | Scientific journals international | STATISTICS |
| Journal of reviews international | Scientific journals international | PUBLISHING AND BOOK TRADE |
| Journal of unconventional theories and research | Scientific Journals International | Multidisciplinary |
| Journal of Sports & Recreation Research and Education | Scientific journals international | Sports education |
| Journal of dissertations | Scientific journals international | HUMANITIES: COMPREHENSIVE WORKS - COMPUTER APPLICATIONS |
| Journal of interdisciplinary & multidisciplinary research | Scientific journals international | Multidisciplinary |
| Journal of economics, banking and finance | Scientific journals international | Economics |
| Journal of international and cross-cultural studies | Scientific journals international | Cultural studies |
| Journal of business and public affairs | Scientific journals international | Business |
| Journal of multicultural, gender and minority studies | Scientific journals international | Sociology |
| Journal of psychiatry, psychology and mental health | Scientific journals international | Psychology |
| Journal of nursing, allied health & health education | Scientific journals international | Nursing |
| Scientific journals international | Scientific journals international | Multidisciplinary |
| Journal of agricultural, food, and environmental sciences | Scientific journals international | Agriculture |
| Journal of creative work | Scientific journals international | |
| Journal of education and human development | Scientific journals international | Education |
| Journal of law, ethics and intellectual property | Scientific journals international | |
| Journal of literature, language and linguistics | Scientific journals international | Linguistics |
| Journal of engineering, computing, and architecture | Scientific journals international | Engineering |
| Journal of medical and biological sciences | Scientific journals international | Medical |
| Journal of humanities and social sciences | Scientific journals international | Social Science and Humanities |
| Open Journal of Hematology | Ross Science Publishers | Medical |
| International Journal of Marketing Practices | Asian Institute of Advance Research and Studies | Economics |
| Genomics and quantitative genetics | Knoblauch Publishing | Biology |
| International Journal of Pharmaceutical Frontier Research | International Journal of Pharmaceutical Frontier Research | Pharmacology |
| Infopreneurship Journal | "This journal is privately published online by Mahmood Khosrowjerdi in Iran." | Economics |
| World of Sciences Journal | Engineers Press | Science and knowledge in general. Organization of intellectual work |
| Indicios (La Rioja) | Universidad Nacional de La Rioja, Instituto Criminalístico de La Rioja | Criminal law |
| Journal of Applied Technology in Environmental Sanitation | Department of Environmental Engineering, Sepuluh Nopember Institute of Technology (ITS) Surabaya, and Indonesian Society of Sanitary and Environmental Engineers (IATPI) Jakarta | Ecology |
| Journal of Applied Phytotechnology in Environmental Sanitation | Department of Environmental Engineering, Sepuluh Nopember Institute of Technology (ITS) Surabaya, and Indonesian Society of Sanitary and Environmental Engineers (IATPI) Jakarta | Ecology |
| Biosciences International | Infofacility | Biological Sciences |
| Journal of Pharmaceutical and Bioanalytical Science | Journal of Pharmaceutical and Bioanalytical Science | Pharmacy |
| Journal of Clinical & Experimental Research | Saujas Publications | Medicine |
| Journal of Current Pharmaceutical Research | Medipoeia | Pharmacy |
| Latin American Journal of Conservation | ProCAT | Zoology |
| Plant Science Feed | Lifescifeed Ventures | Biological Sciences |
| International Journal of Research in Computer Sciences | White Globe Publications | Computer Science |
| Asian journal of pharmaceutical and biological research | Young Pharmaceutical and Biological Scientist Group | Pharmacology |
| Buletin Teknik Elektro dan Informatika | Universitas Ahmad Dahlan | Engineering |
| Applied mathematics in engineering, management and technology | Iran Water and Power Resources Company | Mathematics |
| International Journal of Computer and Distributed Systems | Council for Innovative Research | Computer Science |
| International Journal of Management and Strategy | International Journal of Management and Strategy | Management |
| TheHealth | Lahore Institute of Public Health | Health |
| Amnesia Vivace | Cultural Association Amnesia Vivace | Fine Arts |
| Alfa Redi : Revista de Derecho Informático | Comunidad Alfa-Redi | Law |
| Journal of Chinese Clinical Medicine | Hong Kong Medical Technologies Publisher | Medicine |
| International Journal of Design Computing | University of Sydney | Engineering |
| Polar Bioscience | National Institute of Polar Research | Biology |
| Journal of Multimedia | Academy Publisher | Computer Science |
| Transplantationsmedizin: Organ der Deutschen Transplantationsgesellschaft | Pabst Science Publishers | Internal medicine |
| Wildlife Biology in Practice | Portuguese Wildlife Society | Ecology |
| Great Lakes Geographer | Dept. of Geography, University of Western Ontario | Geography |
| Computer Science Master Research | Universitatea Politehnica din Bucuresti | COMPUTERS |
| Romanian Review of European Governance Studies | Universitatea "Babes-Bolyai" * Centrul "Altiero Spinelli" de Studiere a Organizarii Europene (CASSOE) | PUBLIC ADMINISTRATION |
| International Journal of VLSI and Signal Processing Applications | Innovation Science Publications | COMPUTERS |
| Simbiosis | Universidad de Puerto Rico * Escuela Graduada de Ciencias y Tecnologias de la Informacion | Library and Information Science |
| International Journal of Contemporary Business Studies | Academy of Knowledge Process | BUSINESS AND ECONOMICS |
| Journal of Postcolonial Cultures and Societies | Guild of Independent Scholars | HUMANITIES: COMPREHENSIVE WORKS, SOCIAL SCIENCES: COMPREHENSIVE WORKS |
| E- Journal of Dentistry | E - Journal of Dentistry | MEDICAL SCIENCES - DENTISTRY |
| Journal of Academic and Applied Studies | International Association for Academians | EDUCATION |
| Neues Curriculum | Deutscher Akademischer Austausch Dienst | EDUCATION, HUMANITIES: COMPREHENSIVE WORKS |
| Contemporary Online Language Education Journal | Akdeniz Universitesi | Literature |
| Open Government | Liverpool John Moores University * Faculty of Business & Law | POLITICAL SCIENCE - CIVIL RIGHTS |
| International Journal of Drug Formulation and Research | International Journal of Drug Formulation and Research | Pharmacology |
| European Journal of ePractice | P A U Education | International Law |
| Economics and Finance Review | Global Research Society | Economics |
| Revista Panamericana de Infectologia | Asociacion Panamericana de Infectologia | Public Health |
| Research Center for Educational Technology. Journal | Research Center for Educational Technology, Kent State University | EDUCATION - TEACHING METHODS AND CURRICULUM |
| Molecular and Cellular Pharmacology | LumiText Publishing | Pharmacology |
| NeoAmericanist | University of Western Ontario * Centre for American Studies | HUMANITIES: COMPREHENSIVE WORKS |
| The Electronic Journal of Literacy Through Science | University of California, Davis * School of Education | EDUCATION |
| Calicut Medical Journal | C M C Alumni Association | Medical Sciences |
| Philica | Philica | HUMANITIES: COMPREHENSIVE WORKS, SCIENCES: COMPREHENSIVE WORKS, SOCIAL SCIENCES: COMPREHENSIVE WORKS, TECHNOLOGY: COMPREHENSIVE WORKS |
| Reconstruction | Reconstruction | HUMANITIES: COMPREHENSIVE WORKS |
| Rexter | Centrum pro Bezpecnostni a Strategicka Studia o.s. | Political Science |
MDPI changes the content of a published paper even without any correction note – read yourself

We are completely loosing track if “deemed by the Editorial Office to be a reasonable request” is leading to modifications of text, images or data. And this happened already as I learned recently.
The title is clickbait as nobody really knows what is really relevant in science although some people still think that a few failed postdocs at NSC (Nature, Science, Cell) are the ultimate judges here.
Google also did not know in the 1990ies when they invented the pagerank with the now famous words “The prototype with a full text and hyperlink database of at least 24 million pages is available at http://google.stanford.edu”.
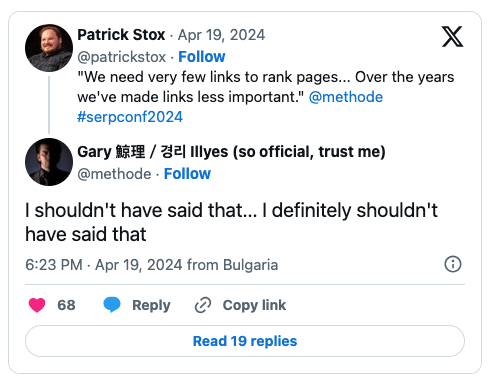
Everybody since then believed in the authoritative power of links but according to Roger Montti they are no more relevant as at a recent conference in Bulgaria Google’s Gary Illyes confirmed that links have lost their importance. And maybe even Google at all?
James Clear adds the reason
Truth and accuracy are not the only things that matter to the human mind. Humans also seem to have a deep desire to belong … Humans are herd animals. We want to fit in, to bond with others, and to earn the respect and approval of our peers. Such inclinations are essential to our survival. For most of our evolutionary history, our ancestors lived in tribes. Becoming separated from the tribe—or worse, being cast out—was a death sentence.” … Convincing someone to change their mind is really the process of convincing them to change their tribe … If you want people to adopt your beliefs, you need to act more like a scout and less like a soldier. At the center of this approach is a question Tiago Forte poses beautifully, “Are you willing to not win in order to keep the conversation going?”
Hard to accept for a scientist but probably true.
“Life is too short to be serious all time”, GILE Journal of Skills Development, Vol. 4 No. 1 (2024)
In this food for thought article, we introduce the ‘Donald Duck Phenomenon’ to consider ten of the more unconventional reasons for publishing in academia. These include
(i) symbolic immortality,
(ii) personal satisfaction,
(iii) a sense of pride,
(iv) serious leisure,
(v) cause credibility,
(vi) altruism,
(vii) collaboration with a friend or family member,
(viii) collaboration with a hero,
(ix) conflict or revenge, and
(x) for amusement.
The article was inspired by the lead author’s social media search for a co-author with the surname ‘Duck’. Through LinkedIn, the lead author, Associate Professor William E. Donald, who is based in the UK and specialises in Sustainable Careers and Human Resource Management, found a collaborator, Dr Nicholas Duck, who is based in Australia and specialises in Organisational Psychology. While the collaboration may appear to be somewhat ‘quackers’, per one of Donald Duck’s famous phrases “Life is too short to be serious all the time, so if you can’t laugh at yourself then call me… I’ll laugh at you, for you”. We hope that this article offers some interesting insights and acts as a way to stimulate conversation around unconventional reasons for publishing in academia.
At this point, I feel bleak at the prospect of typing them out again. The problems with overpublication, ‘publish or perish’ culture, abusive lab environments, analytical flexibility, p-hacking, clinical trial registration games, grant front-running, intellectual capture, nonsense journals, fake journals, peer review manipulation, moral entrepreneurship, etc. precede the present discussions of paper mills and active falsification/fabrication cases…
I have tried at least four times in my memory to write out and codify how I would start an institute to combat these problems. Specifically, a formal organization under a 501c3 structure designed to address the problem.
In Germany we have the IQWIQ, an independent Institute for Quality and Efficiency in Health Care who examines the benefits and harms of medical interventions for patients. But they don’t care about all the medical nonsense studies around. And without PubPeer we wouldn’t even know the nonsense…
Create a PHP script that can read a CSV in the form start_date, end_date, event and output as ICS file
function convertDate($date)
{
$dateTime = DateTime::createFromFormat('m/d/Y', $date);
if ($dateTime === false) {
return false; // Return false if date parsing fails
}
return $dateTime->format('Ymd');
}
// Function to escape special characters in text
function escapeText($text)
{
return str_replace(["\n", "\r", ",", ";"], ['\n', '\r', '\,', '\;'], $text);
}
// Read CSV file
$csvFile = 'uci.csv'; // Replace with your CSV file name
$icsFile = 'uci.ics'; // Output ICS file name
$handle = fopen($csvFile, 'r');
if ($handle !== false) {
// Open ICS file for writing
$icsHandle = fopen($icsFile, 'w');
// Write ICS header
fwrite($icsHandle, "BEGIN:VCALENDAR\r\n");
fwrite($icsHandle, "VERSION:2.0\r\n");
fwrite($icsHandle, "PRODID:-//Your Company//NONSGML Event Calendar//EN\r\n");
// Read CSV line by line
while (($data = fgetcsv($handle, 1000, ',')) !== false) {
$startDate = convertDate($data[0]);
$endDate = convertDate($data[1]);
print_r($data) . PHP_EOL;
echo $startDate;
if ($startDate === false || $endDate === false) {
continue;
}
$event = escapeText($data[2]);
// Write event to ICS file
fwrite($icsHandle, "BEGIN:VEVENT\r\n");
fwrite($icsHandle, "UID:" . uniqid() . "\r\n"); // Unique identifier
fwrite($icsHandle, "DTSTART;VALUE=DATE:" . $startDate . "\r\n");
fwrite($icsHandle, "DTEND;VALUE=DATE:" . $endDate . "\r\n");
fwrite($icsHandle, "SUMMARY:" . $event . "\r\n");
fwrite($icsHandle, "DESCRIPTION:" . $event . "\r\n");
fwrite($icsHandle, "END:VEVENT\r\n");
}
// Write ICS footer
fwrite($icsHandle, "END:VCALENDAR\r\n");
// Close files
fclose($icsHandle);
fclose($handle);
echo "ICS file generated successfully.";
} else {
echo "Error: Unable to open CSV file.";
}
Source data are from UCI and output is here from where it can be added as a calendar. BTW created also my first “hello world” Swift/iPhone app using this source although this took a bit more time…
Give me now the flute then, and sing
Disease and drug, forget them
for people are lines written
but with water
Lior Pachter has an interesting observation
which goes back to an old article of Richard Guy extracting four major issues in interpreting data
Unfortunately this seems to describe the way we think and even worse – this is what the science system promotes: the spectacular, the unexpected, the fascinating news.
To continue his story, what is the lifetime of the spurious idea? In many instances effects are declining rapidly for example in intelligence research. It took me some time to find the first paper that I remember – it was in 2001 that John & Despina wrote that the results of the first study correlate only modestly with subsequent research on the same association. This was confirmed in 2005
Of 49 highly cited original clinical research studies, 45 claimed that the intervention was effective. Of these, 7 (16%) were contradicted by subsequent studies, 7 others (16%) had found effects that were stronger than those of subsequent studies, 20 (44%) were replicated, and 11 (24%) remained largely unchallenged.
A scandal? The list of failed studies is long, including all areas of biomedicine already back in 2015.
While AI may not break science for being backwards directed, there are already the first companies doing AI interviews.
Could become a big problem whenever universities are also using this type of job interviews.
Just like journals who use AI for peer review
The authors of the study1, posted on the arXiv preprint server on 11 March, examined the extent to which AI chatbots could have modified the peer reviews of conference proceedings submitted to four major computer-science meetings since the release of ChatGPT. Their analysis suggests that up to 17% of the peer-review reports have been substantially modified by chatbots — although it’s unclear whether researchers used the tools to construct reviews from scratch or just to edit and improve written drafts.
Es tut der Wissenschaft nicht gut, wenn man probiert, sie auf politische Ziele festzulegen, selbst wenn diese weithin gesellschaftlich akzeptiert sind. Was ist die Alternative? Eine altmodische Idee von Max Weber. Sie heißt: Werturteilsfreiheit. Damit wollte Weber die Sozialwissenschaften gegen eine Vereinnahmung durch links und rechts bewahren. Wissenschaftler, so Weber, sollen erforschen, wie die Welt ist, nicht ihre Autorität nutzen, um anderen einzureden, wie die Welt sein sollte. Denn wo sich Werte widersprechen, kann man nicht wissenschaftlich entscheiden, welche richtiger sind. Forscherinnen und Forscher sollten sich deswegen aus politischen Diskussionen fernhalten.
oh ja, das hatte ich auch einmal im Ärzteblatt geschrieben was ich denn von Umweltepidemiologie halte, die vor 30 Jahren gegen und nun im Mainstream Nonsense Ergebnisse produziert.
Und nun auch in der neuesten ZEIT “warum eine Universität überhaupt eine politische Haltung hat”.

Also Positivismusstreit reloaded?
Nein, bestimmt nicht. Ohne einzelne Werturteile geht es natürlich nicht, sie sollten im Zweifel aber als “Conflicts of Interests” am Ende jedes wissenschaftlichen Artikels stehen. Wo die Tatsachen enden und wo die Interpretation anfängt.