When getting important documents for review I usually check them for plagiarism. One of the best address seems docoloc – try it out, they have Continue reading More on plagiarism
All posts by admin
Let us build a city and a tower, whose top may reach unto heaven
The great pyramid of Giza by Cheops (Khufu, ΧÎωψ) is a true mystery. SPIEGEL online now reports another attempt to explain how the pyramid has been constructed. A French architect believes Continue reading Let us build a city and a tower, whose top may reach unto heaven
The winner’s curse
is another attempt to explain why replication fails frequently in genetic epidemiology. Zöllner and Pritchard write in the AJHG (their server is currently down)
For a variant that is genuinely—but weakly—associated with disease, there may be only low or moderate power to detect association. Hence, when there is a significant result, it may imply that the genotype counts of cases and controls are more different from each other than expected. Consequently, the estimates of effect size are biased upward. This effect, which is an example of the “winner’s curse” from economics depends strongly on the power of the initial test for association. If the power is high, most random draws from the distribution of genotype counts will result in a significant test for association; thus, the ascertainment effect is small. On the other hand, if the power is low, conditioning on a successful association scan will result in a big ascertainment effect.
I haven´t fully understood the following argumentation, but promise to revisit it some times later, yea, yea.
More statistics
indexed.blogspot.com has many interesting data visualizations – I like them!
A more serious approach for interpreting health statistics can be found in the Lancet. Continue reading More statistics
Gary Taube’s limits and my interest in molecular epidemiology
Curative medicine contributes only 10% to 40% to individual health (figures are depending on models and methodology according to a recent commentary in the Deutsche Ärzteblatt, for milestones check the BMJ) – a reason why I finally decided to become an epidemiologist. Continue reading Gary Taube’s limits and my interest in molecular epidemiology
Divide and conquer
Science has an interesting paper that relates to an earlier post here and a central question in immunology. How do T cells differentiate int both short-lived effector cells (that combat infections) and long-lived memory cells. Continue reading Divide and conquer
Anticipating trouble
Science magazine today reports another ego trip.
A U.S. company [454] has begun to trickle out information on a unique DNA study it calls “Project Jim,†a crash effort to sequence the entire genome of a single individual. The results are likely to be made public this summer. Anonymity is out of the question: It has already been announced that the genome belongs to James D. Watson, winner of the Nobel Prize and co-discoverer of DNA’s structure. Watson won’t be alone: Harvard Medical School has approved a plan by computational geneticist George Church to sequence and make public the genomes of well-informed volunteers—including his own. And J. Craig Venter says his nonprofit institute will soon release a complete version of his genome.
My daily newsletter says that Roche is going to acquire 454 for $155M and plans to use the sequencer for IVD applications, I hope they will forget “Project Jim” somewhere on a harddisk.
Antedisciplinary Science
.. another thoughtful essay by Sean Eddy in PLOS Computational Biology cites the NIH Roadmap Initiative
The scale and complexity of today’s biomedical research problems demand that scientists move beyond the confines of their individual disciplines and explore new organizational models for team science. Advances in molecular imaging, for example, require collaborations among diverse groups—radiologists, cell biologists, physicists, and computer programmers.
which sounds great like all interdisciplinary science but has also all the drawbacks (“to temper the wind to the shorn lamb” seems to be the English translation of the German “weakest ring of the chain”).
Progress is driven by new scientific questions, which demand new ways of thinking. You want to go where a question takes you, not where your training left you.
Sure, the game is more about interdisciplinary people than interdisciplinary teams
A motley crew of misfits
and not EU accountants drive progress.
How much do you think a scientific blog post is worth
(in US dollars) asks Pimm – the partial immortalization blog. A first response to this question -based on Google adsense revenues- is about $0.47/post. I think that prices depend on context – from negative balance (wasted time) to a new research direction (+tenure +$100,000) there is everything possible.
Collins: Testimony on the threat of genetic discrimination
The testimony of Francis Collins before the subcommittee on Health is now online
A recent NIH study of families at risk for hereditary nonpolyposis colorectal cancer … revealed that the number concern expressed by participants regarding genetic testing was about losing health insurance, should the knowledge of their genetic test result be divulged or fall into the “wrong hands” … Unless Americans are convinced that their genetic information will not be used against them, the era of personlized medicine may never come to pass. The rest would be a continuation of the current one-size-fits-all medicine, ignoring the abundant scientific evidence that the genetic differences among people help explain why some of us benefit from a therapy while others do not.
Autosomal inheritance of sex-linked marker
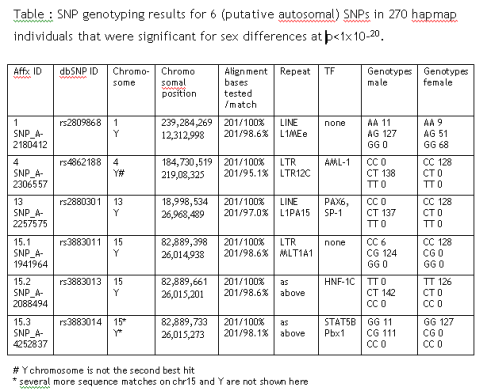
While optimizing the analysis strategy for a 500,000 SNP Affymetrix array set, I found 6 autosomal SNPs that show highly significant sex-dependent allele differences: rs2809868, rs4862188, rs2880301, rs3883011, rs3883013 and rs3883014.
Sure, there could be autosomal marker that influences male/female outcome but there is a more likely explanation: All SNPs have paralogue sequence stretches on the Y chromosome that are co-amplified during PCR. From the initial genotyping results it is most likely that only the Y chromosomal stretch is being mutated in SNP 4, 13 and 15.2.
These SNPs are perfect sex marker, as they include an autosomal control allele (in comparison to pure Y markers like SNPs in SRY). They are always unambiguous (in contrast to pure X marker where only heterocygotes are informative).
They even offer advantage to commercial STR kits of the Amelogenin/Amely gene situated (in the Y parautosomal region) as they would not be affected by excess homologous X chromosomal material as often found in forensic situations. In addition, they might overcome some other weakness of the Amelogenin test where a second assay is usually recommended.
If you will ever see a case-control study that is highlighting any of these SNPs, you can be sure that this study had a distorted male-female ratio between case and controls.
The other way around
One of my favorite pictures is the Orangenesser by Baselitz – his speciality is to paint his subjects the other way around to free the subject from its content (this is at least what art curators say).
DNA inversion rearrangements seem to be also more frequent in the human genome as previously thought, see the following PNAS preprint:
Three such regions … on chromosomes 3, 15, and 19, were analyzed. … The results obtained indicate that recurrent genomic rearrangements occur at relatively high frequency in somatic cells. Interestingly, the rearrangements studied were significantly more abundant in adults than in newborn individuals, suggesting that such DNA rearrangements might start to appear during embryogenesis or fetal life and continue to accumulate after birth.
More paranormal inheritance
A funny study in PNAS is broadening our view of paranormal inheritance by showing germline chimerism in marmosets:
Using microsatellite DNA markers, we show here that chimerism in marmoset (Callithrix kuhlii) twin … somatic tissue .. found to be chimeric. Notably, chimerism was demonstrated to be present in germ-line tissues, an event never before documented as naturally occurring in a primate. In fact, we found that chimeric marmosets often transmit sibling alleles acquired in utero to their own offspring.
Bad news for all who believe in human paternity testing and 99,999999% probability – and more strange stories (semi-identical twins discovered – hermaphrodite reveals previously unknown type of twinning) at the Nature news blog.
Science dresscode
Sciencesque has noticed it first: T shirts at Biomed Central for sale. Do your really want that shirt?
Another path from vitamin D to allergy
As far as I know vitamin D3 influences only RUNX2 expression (RUNX2 has a VDRE, possibly also RUNX3 but not RUNX1?). The RUNX factors
colocalized in common subnuclear foci. Furthermore, RUNX subnuclear foci contain the co-regulatory protein CBFβ, which heterodimerizes with RUNX factors, and nascent transcripts as shown by BrUTP incorporation. These results suggest that RUNX subnuclear foci may represent sites of transcription containing multi-subunit transcription factor complexes.
Variants in RUNX1 have already been reported earlier to be weakly associated with IgE serum levels in Korea.
A Nature paper todays reports that Foxp3 controls Treg function by interacting with AML1/RUNX1 – is there any connecting path?