This content has restricted access, please type the password below and get access.
Category Archives: Allergy
The logo site
Sequence logos are fun: www-lmmb.ncifcrf.gov is the ultimate site for many demo figures. It provides even a link where you can manufacture your own logo. Here is mine which is derived from a TCR consensus sequence:
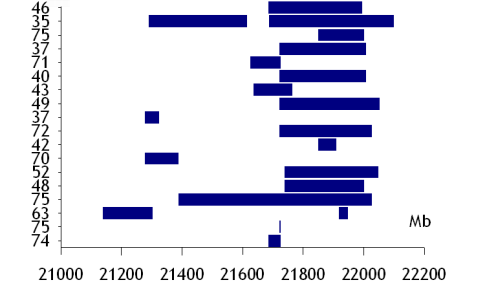
Genome plots with Sigmaplot
Here are some instructions how to create genome coordinate plots with Sigmaplot. The Systat Sigmaplot FAQ recommends for this kind of figures “floating bars” that
can be created using two different mechanisms. If you just need a simple floating bar chart, you can use a box plot of two values per column. Enter the top and bottom ranges for your bars in each column, and then create a box plot…
If you need to create grouped, floating bar charts, you will need … a masking plot. Create a grouped bar chart, using the upper values of the bars. Create a second bar chart for the same graph (click the graph, then click the grouped bar chart icon from the Graph toolbar), this time, using the lower values of the bars.
None of these methods works with fill patterns. Also repeated segments (at the same level) are not possible.
So I have worked out therefore another method using a stacked bar plot:
(1) put in the first column an increasing value 1 … 10 (->gets Y axis)
(2) put into the second column zeros (->gets start set 1)
(3) third colum: first feature start (->gets end set 1)
(4) fourth column: zeros (->gets start set 2)
(5) fifth column: first feature end (->gets end set 2)
(6) and so on
(7) assign colors: white for start set 1, blue for start set 2, white for start set 3 and so on
You should then end up with a plot like this
“Science advances in stages and no story is complete”
Sometimes we think we are alone with our difficulty of getting a good paper published. In lucid moments, however, we find others making the same experience- see a commentary in Science
The stress associated with publishing experimental results–a process that can take as long as obtaining the results in the first place–can drain much of the joy from practicing science Continue reading “Science advances in stages and no story is complete”
Fabrics for atopic dermatitis
Most of the medical research into causes of allergy is more or less irrelevant to allergy sufferers. Here is an exception [author] if you are suffering from atopic dermatitis [paper] Continue reading Fabrics for atopic dermatitis
Probability that a genetic association is false positive
An (anonymous) reviewer of our forthcoming EJHG paper on IgE and STAT3 pointed me towards a JNCI paper that has a nice supplement – an excel sheet to calculate the probability that a positive report is false. It basically relies on (i) the magnitude of the p-value (ii) statistical power and (iii) fraction of tested hypothesis. While we certainly know (i) and (ii), (iii) is always hard to know with many datasets including hundreds of traits that allow indefinite numbers of subgroups. Are you really interested in a new paper about “An African-specific functional polymorphism in KCNMB1 shows sex-specific association with asthma severity” that encompasses 1 of virtually 100 ethnic groups; 1 or virtually 25000 genes; 1 of 2 sexes; 1 or virtually 50 asthma related traits, yea, yea.
View plink GWA results as genome browser track
This R script will parse plink‘s GC adjusted output to a GFF file that can be uploaded as genome browser track file. Just adjust the pathname at the beginning…
1: | |
Will the Data Deluge Make the Scientific Method Obsolete?
A new Edge article answers this question. According to Chris Anderson, we are at “the end of science”, that is, science as we know it.
The quest for knowledge used to begin with grand theories. Now it begins with massive amounts of data. Welcome to the Petabyte Age.
Yesterday I reviewed a paper that crunches massive amount of data (and even found a new pathway for asthma). Nevertheless I was asking the question if this wishful thinking? Just take the next gene in one region and the overnext in another one and I would come up with a completely different pathway. This is all about association and not by the traditional “theorize, model, test it” way of science we have been brought along, yea, yea.
Asthma and allergy trend
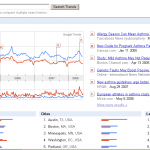 Besides traditional and expensive epidemiological studies to compare disease prevalence over time, the net offers also some nice research tools – click on the thumbnail to see Google asthma trend results by today. The query counts are largely consistent with most recent scientific papers seeing the asthma epidemic now at a (high) plateau. Did the environmental risk now saturate all individuals at risk?
Besides traditional and expensive epidemiological studies to compare disease prevalence over time, the net offers also some nice research tools – click on the thumbnail to see Google asthma trend results by today. The query counts are largely consistent with most recent scientific papers seeing the asthma epidemic now at a (high) plateau. Did the environmental risk now saturate all individuals at risk?
The many problems of GWAs
genetic-future has an excellent article why the recent genome scans failed (i) alleles with small effects? (ii) population differences? (iii) epistatic interactions? (iv) cnvs more relevant than snps? (v) epigenetic inheritance? (vi) disease heterogeneity? It is a thorough review better than everything Continue reading The many problems of GWAs
Vitamin D: A 18%! reduction in early infant mortality
A study published earlier this year in the Lancet found a 18% reduction in mortality when women obtained supplements during pregnancy until 90 days post partum including additional 800 ug retinol, 200 IU vitamin D, 10 mg vitamin E, 70 mg ascorbic acid, 1.4 mg vitamin B1, 18 mg niacin, 1.9 mg vitamin B6, 2.6 ug vitamin B12, 15 mg zinc, 2 mg copper, 65 ug selenium and 150 ug iodine. Much of the effect will be due to the vitamin D supplementation Continue reading Vitamin D: A 18%! reduction in early infant mortality
Not all that wheezes is asthma
A new abstract at the recent ATS congress now clears the 2007 controversy between the Camargo and Gale studies on the effect of vitamin D: Wheezing is not asthma (as the atopy component is missing?). Continue reading Not all that wheezes is asthma
CF and PHP at the same time
For a clinical research project I need a fast webserver that understands both COLD FUSION (for the existing database stuff) and PHP (for dokuwiki). As this is a non-commercial project, I decided for the RAILO 2.0 community version that works right out of the box as it includes the Resin webserver and the H2 database as well. Following some recommendations found on the net, php is running now with with the fastcgi option showing good acceptable performance with eAccelerator.
1: | |
CD 14 now also on the vitamin+allergy list
Just for curiosity I am collecting a list of allergy genes that are vitamin D dependent. The list is already rather long but now there is a prominent addition: CD14. Known as asthma gene for many years the vitamin D dependency isn’t such clear. A clever analysis, however, now shows that there is an intermediate step involved Continue reading CD 14 now also on the vitamin+allergy list
Vitamin D as an adjuvans to specific immunotherapy
Despite the known allergy promoting effect of vitamin D in early childhood, there is mounting evidence that it may have beneficial effects during specific immunotherapy. A paper on “IL-10-inducing adjuvants enhance sublingual immunotherapy efficacy in a murine asthma model” by researchers from a French allergen company Continue reading Vitamin D as an adjuvans to specific immunotherapy