Von den Ereignissen in der 1986 geschlossenen Asthma-Kinderheilstätte in Bad Reichenhall höre ich heute morgen zum ersten Mal in einem Podcast von BR24. Der Missbrauch geht dabei weit über die unsäglichen Verschickungsheime der 50er und 60er Jahre hinaus, die für Ihre Erziehungsmethoden berüchtigt waren. Continue reading Die Asthma-Kinderheilstätte Bad Reichenhall
Tag Archives: asthma
An update on the asthma exome
Here is a quick update on some genes of my recent asthma exome paper coming now from the 1 M exome paper published yesterday as a preprint.
Also ClinVar shows that the IL33 receptor is not “essential” making anti IL33 receptor antibodies like etokimab, itepekimab, tozorakimab a safe therapy although not being effective in any LOF mutation carrier.
The most interesting thing in the preprint is in supplemental table 2 with the s-het values for 16,704 genes. From that table I have selected my favorite target IL33 receptor together with TLR1, ALOX15, GSDMA, IL13 and IKZF3 ( BTNL2 could not be found in the list).
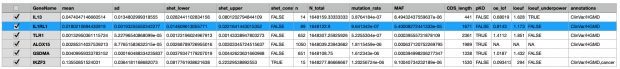
IKZF3 would be dangerous to be touched (see my 2008 commentary) while in the 2022 exome paper I also found only protective variants in the 5′-UTR but not any LOF variant – probably as IKZF3 is the only essential gene in the list.
So what’s next? I am still thinking how to reduce my exome set to the causal variants as half of the mutations are probably LD artefacts. And well, it would be super interesting to examine now two extreme inbred populations for their mutation spectrum, loosing either asthma variants by healthy (Amish) or diseased founders (Tristan da Cunha). Unfortunately there is little hope that this will happen – current science is built more on competition than collaboration.
The Lancet and scientific integrity
We have learned in the past that the Lancet published editorials that clearly separated the journal from the publisher Elsevier
Reed Elsevier’s response is that the sale of military equipment is legal, government supported, and tightly regulated. However, The Lancet‘s collaborations in child survival and health-systems strengthening, for example, risk being tainted by Reed Elsevier’s promotion of the “selling process” of arms.
Of course you can’t sell weapons and distance yourself from selling weapons at the same time…
What did Sequana really find at Tristan da Cunha?
The Tristan da Cunha asthma study was leading to patent US6087485A. The patent describes
highly significant linkage in the genome scan (p=0.0001 for history of asthma and p=0.0009 for methacholine challenge) … at D11S907, a marker on the short arm of chromosome 11.
D11S907 or AFM109YA1 is a microsatellite marker located at 11p13 in a gene known as EHF (ETS homologous factor). There should be 2 genes in close proximity of the marker: ASTH1I and ASTH1J.
ASTH1I and ASTH1J were detected by exon trapping. ASTH1I exons detected a 2.8 kb mRNA expressed at high levels in trachea and prostate, and at lower levels in lung and kidney …
ASTH1J exons detected a 6.0 kb mRNA expressed at high levels in the trachea, prostate and pancreas and at lower levels in colon, small intestine, lung and stomach.
The sequences of table 2 in the patent are sufficient now to locate ASTH1I and ASTH1J. Continue reading What did Sequana really find at Tristan da Cunha?
Censored IL33 research
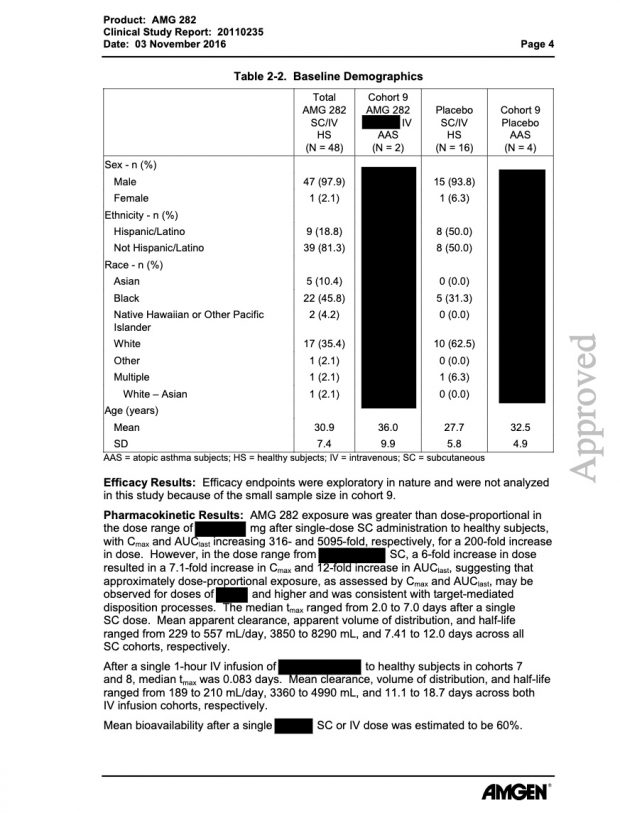
the AMGEN website …
Rare variants in GSDMA/B /IKZF3 but not in ORMDL3 are relevant for asthma
It was a heated debate (paper, blog) that was open until a most recent study that I am depicting here in more detail.
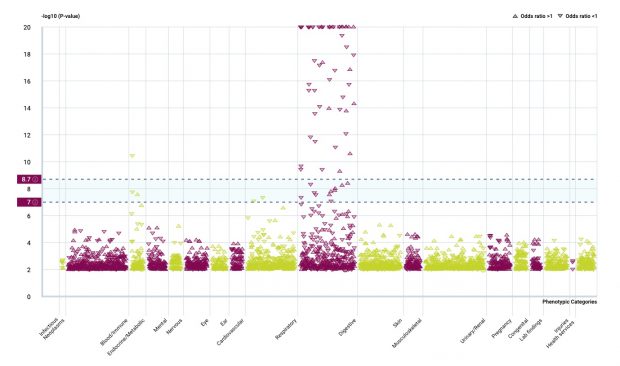
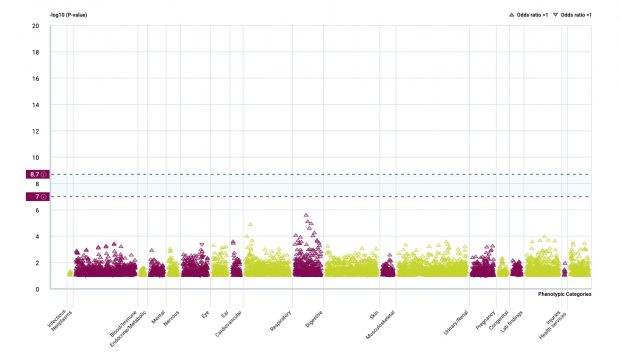
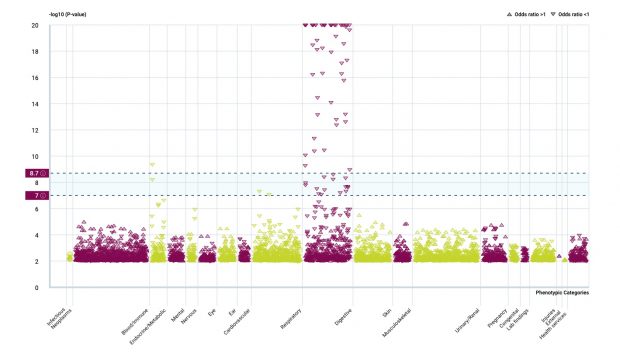
Maybe there is an interaction by methylation?
The analysis of SNP effects is ongoing while it now looks like ORMDL3 SNPs were just hitchhikers, at least what I see.
Addendum 4.9.2023
Maybe there is even a vitamin interaction?
How atopic dermatitis is linked to vitamin D and how IL33 splice variants associate to eosinophil numbers
(first published 12 Dec2020 and revised 10Dec2021)
We had a major discussion right before our 2010 paper where I argued that rare variants should have been included into our asthma/allergy/dermatitis GWAS. Ten years after there is now a nice paper using massive exome sequencing that finally includes them.
It seems that the respiratory tract isn’t so much influenced by rare gene variants but that there is a strong effect in the immune system.
And there is another interesting fact.
…Surveying the contribution of rare variants to the genetic architecture of human disease through exome sequencing of 177,882 UK Biobank participants …if we look at the …. European population who are carriers of a filaggrin (FLG) PTV, we find those carriers have significantly higher risk for well-known associations, such as dermatitis … and asthma … Concomitant increases in vitamin D levels suggest … increased sensitivity to ultraviolet B radiation.
So far, I have only assumed an asthma/allergy priming effect of oral vitamin D in the newborn gut. This paper now argues for an increased vitamin D sensitivity also in the skin of FLG dermatitis patients which is interesting given the largely contradictory data of serum vitamin D and atopic dermatitis. Maybe dermatologists should focus their research more on skin and local vitamin D turnover?
-II-
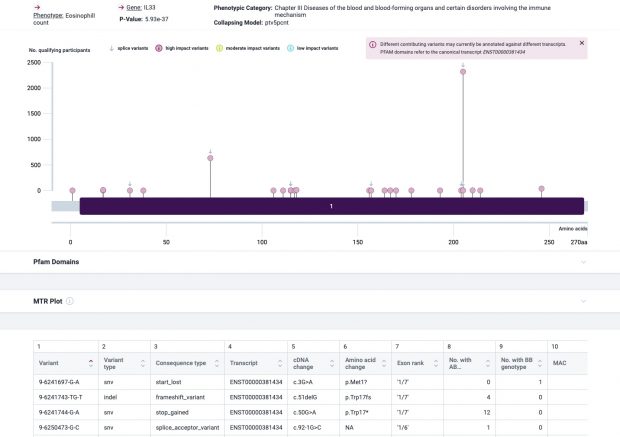
The most prominent IL33 variant carried by over 2,300 people is splice acceptor 9-6250473-G-C followed by 600+ individuals with splice donor 9-6250600-G-T.
There are not too many carriers of this variant by the sheer amount of 177,882 participants. We nevertheless know already something about the seven IL33 splice variants since 2012.
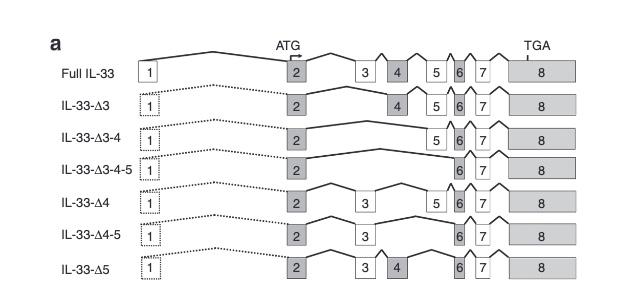
with updates in 2016
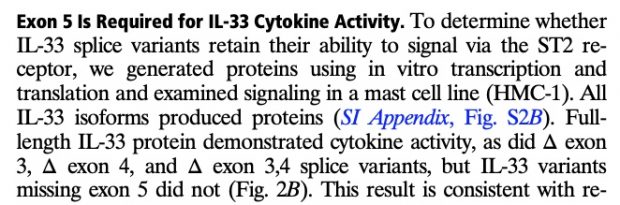
as well as in 2017
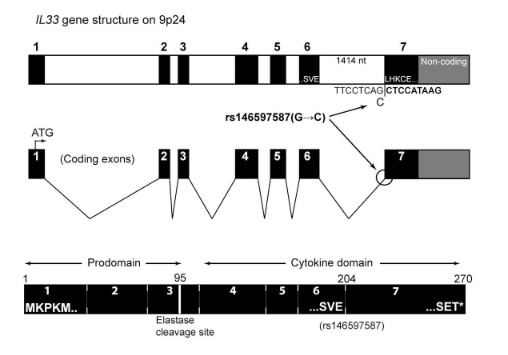
So I did a sequence match to compare the new finding with these older publications.
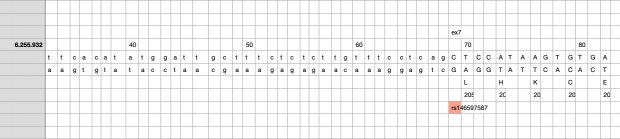
Indeed, the 2017 paper already described rs146597587 which is probably identical to the splice acceptor 9-6250473-G-C in Astra UK Phewas (genome positions do not match – I used hg19 while I don’t know the Astra reference) . Astra says also c.613-1G>C while rs146597587 is just upfront of my codon 205 (3*205=615) whatever that means.
The Astra UK Phewas at least confirms the Iceland paper above
rs146597587-C associates with lower eosinophil counts (ß= -0.21 SD, P = 2.5×10-16, N = 103,104), and reduced risk of asthma in Europeans (OR = 0.47; 95%CI: 0.32, 0.70, P = 1.8×10-4, N cases = 6,465, N controls = 302,977). Heterozygotes have about 40% lower total IL33 mRNA expression than non-carriers and allele-specific analysis based on RNA sequencing and phased genotypes shows that only 20% of the total expression is from the mutated chromosome. In half of those transcripts the mutation causes retention of the last intron, predicted to result in a premature stop codon that leads to truncation of 66 amino acids.
So it is basically a rediscovery meaning that we reached saturation.
Update on asthma and allergy genes
Even the most recent attempt by Open Targets integrating basically everything
GWAS Catalog and UK Biobank) with transcriptomic, proteomic and epigenomic data, including systematic disease–disease and disease–molecular trait colocalization results across 92 cell types and tissues … trained a machine-learning model … to distinguish causal genes from neighboring genes, outperforming a naive distance-based model
ended up with nothing new
I am glad to see, however, that my criticism of the ORMDL3 hype has been taken up by other authors as well
In the first report of the association of the 17q21 locus with asthma, Moffatt et al. suggested ORMDL3 as a promising candidate on the basis of gene expression studies in EBV-transformed lymphoblastoid cell lines 5. However, the function of ORMDL3 remains to be fully elucidated and it is possible that other genes in this region, or more distant genes, contain the true causal variants.
while ORMDL3 isn’t appearing at all in the above analysis. So ORMDL3 seems to suffer the same fate as FcER1b/MS4A2 identified by the same group.
Vitamin D Kids Asthma Study – Criticism is not justified
On Aug 12, Science magazine published a strange journalistic article of an ongoing vitamin D asthma study. I responded on the next day
https://twitter.com/science_surf/status/1426145394518630400
while now also the former editor-in-chief of Science Jeremy Berg noted
This is some of the most irresponsible and intellectually lazy science journalism I have seen from a venue like @sciencemagazine that should, indeed must, do better
— Jeremy Berg (@jeremymberg) October 27, 2021
Science printed the rebuttal of Berg
Piller minimizes the rationale used to select the placebo-controlled trial design and suggests that there is agreement that such a design is unethical… The News story notes that the majority of children in the trial were Black and states that this constitutes overrepresentation… Rather than being criticized, this trial should be commended for inclusion of appropriate trial participants…. Piller writes that participants were at increased risk of fractures and that the nine bone fractures experienced by study participants were more than anticipated, without specifying the magnitude of any increased risk or the anticipated number of fractures. However, there is no consensus that any increased risk exists…
Piller misrepresents the Vitamin D Kids Asthma Study (“Vit-D-Kids” or “VDKA”) . He reports concerns about the study’s design, participant safety and selection, consent forms, and report trans-parency. These doubts are unfounded. VDKA ethically investigated a potentially important treatment for childhood asthma.
Unfortunately the response of the news editor Tim Appenzeller is so weak that I would recommend to search now not only for a new correspondent but also a new editor.
Missing heritability
It is always nice if you are being cited after some years ;-)
There is an important article that I have been reading this morning “Missing heritability of complex diseases: case solved?” Emmanuelle Génin is correct by questioning the statistical models underlying our heritability measures.
It is therefore possible to directly quantify the contribution of these genetic variants to phenotypic variance and measure what is called “genomic heritability”. Briefly, the idea is to infer the proportion of variance that can be explained by a linear regression on a set of markers used as explanatory variables.
This genomic heritability seems to be in fact a poor mathematical representation of a biological trait. >1000 hits in >500.000 individuals, does that make any sense at all in terms of biology?
More recently, it was proposed that rather than polygenic, the genetic architecture of common diseases could in fact be “omnigenic” with mainly all active genes affecting every complex traits.
which is fatalistic but justified IMHO. But there could be solutions. We proposed undetected rare variants
When adding these rare and low-frequency variants to the common variants associated with height, 27.4% of the variation can be explained.
while the results are not impressive. We proposed also structural variants (CNVs), but again no convincing effect so far. Allele dosage? Disappointing . GxE, interaction with genome background? Difficult, highly complex. Epigenetic environmental influences ? Sure but where to start?
The “GIGO (Garbage-In Garbage-Out) syndrome in genetics” as coined by Emmanuelle Génin is one of the main reasons why I mostly dropped out of the field.
We believe that the “missing heritability problem” is an ill-posed problem. Solving it by refining over and over again statistical models derived under the “polygenic paradigm” or using more and more sophisticated sequencing machines will not answer the fundamental and biological question of why that individual is affected by the disease and that other individual is not.
My last sentence would be: Phenotypic correlations between relatives determine heritability. So we should go back to family studies if we want to explain heritability.
Hope dies last
Maybe the Nature editors should have read their own writings as the “roadmap for regulation” (19 Feb 2015) states correctly about gene methylation Continue reading Hope dies last
History of allergy research In Germany
Using data described in Science Magazine (basically 4% of all published books) I constructed a plot showing an update to the 100 year old mystery why we get allergic to certain substances.
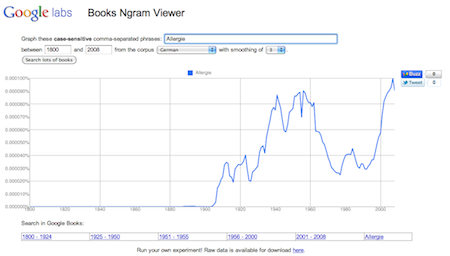
There seems to be a good chance for 2011 if the line will further increase…
The largest study so far on serum cytokines
We just published the largest study so far of human serum cytokines providing for the first time reference values.
In this study we investigated serum samples from 944 individuals of 218 asthma-affected families by a multiplex, microsphere based system detecting at high sensitivity eleven asthma associated mediators: eotaxin (CCL11), granulocyte macrophage stimulating factor (GM-CSF), interferon gamma (IFNγ), interleukin-4 (IL-4), IL-5, IL-8, IL-10, IL-12 (p40), IL-13, IL-17 and tumor necrosis factor alpha (TNFα). Continue reading The largest study so far on serum cytokines
The asthma and glaucoma island
Dr. Zamel, one of the PIs of the Tristan da Cunha study pointed me today to the interesting 30 min BBC documentation online at Allergy Canada
Allergy Island is an exclusive documentary on the history of asthma in Tristan da Cunha and the discovery of the gene related to asthma in that highly inbred community by the expedition that I did in 1993. I went in May 2008 to Tristan da Cunha with the BBC crew to film both parts for the entire month.
Factory science
There seems to be a GWAS repository that has an entry of an asthma study otherwise not known in the biomedical literature and – as see on the screenshot -results tables are empty. Continue reading Factory science