Usually “materials and methods” section is in the second paragraph; some journals put it also at the end of a paper. As a reviewer I have always insisted that this heading should be extended to “Patients, materials and methods” while in epidemiology we frequently use the term subjects (BTW epidemiologists have a rather militaristic vocabulary: recruited, cohorts :-) ). The anonymous reviewer of a previous paper now pointed out: Use the phrase “participant” throughout and not “subjects” which has reductionist connotations. I promise to use “participants” from now on, yea, yea.
Not an academic project, not an industry project
Not an academic project, not an industry project, but still earn money with knowledge? Innocentive posted the request #4470259 for an ALS biomarker. Deadline is Nov 06, 2008 and you will get $1,000,000 USD for problem solving. Yea, yea.
Peer production
firstmonday has an interesting article about the limits of self-organization and “laws of quality”. Given 52 million tracks in the Gracenote database, 1 million entries in Wikipedia and 17,000 books in project Gutenberg, Paul Duguid throughly examines the two laws of quality
- Linus law: “given enough eyeballs, all bugs are shallow” which means that almost every error will be discovered and ultimately fixed
- Graham law: “people just produce whatever they want; the good stuff spreads, and the bad gets ignored”
Although more professionalized, similar principles operate in science. With these large genetic studies, I have the feeling that most errors occur at the interfaces, during hand-shaking of disciplines. There are certainly only a few people that can design a study, examine a patient, go to the laboratory, analyze and annotate the data and publish them. This means that even many eyeballs can not look around the corner and that it will take many years for the “good stuff to spread”. Yea, yea.
Pharmacogenetic tests on the market
Certainly one of the best web resources for pharmacogenetics is the PharmGkB database that collects all kind of data about the relationships among drugs, diseases and genes. Of course you could sequence your genome or run expression profiling on a liver sample. However, you are probably here to find out what (serious!?) pharmacogenetic tests are already on the market.
Much can be said about the usefulness of such tests; I have doubts if there will ever be such personalized treatment as I can foresee some logistic problems to validate it ;-) More likely are group based therapies, maybe restricted to geographic ancestry. Here is a (first and very) preliminary collection of commercially available pharmacogenetic tests:
- CYP2D6, CYP2C9 and CYP2C19 collectively account for about 40 percent of drug metabolism mediated by cytochrome P-450 (Roche). The AmpliChip CYP450 Test is the world’s first pharmacogenetic microarray-based test approved for clinical use. CYP2D6 metabolizes codeine into morphine. A variation in CYP2D6 varies with race and leads to a lower elimination rate of the antidepressants Prozac (a selective serotonin reuptake inhibitors); the alternatively used drug Celexa is metabolized by CYP2C19 (as well as omeprazole). Other examples include clopidogrel (metabolized by CYP3A4) and cyclophosphamide (by CYP2B6) and vitamin K (by CYP2C9)
- NAT2*5A, NAT2*6A, NAT2*7A/B and NAT2*14A carriers are rapid and slow acetylators for example of isoniazid or procainamide (Roche)
- HER2+ women may get herceptin (Roche, Bayer, PathVysion)
- TP, DPD are the rate limiting catabolic enzymes of 5-fluorouracil metabolism (Roche)
- Mitochondrial A155G variants are tested for aminogylcoside side effect (Humatrix)
- A Warfarin sensitivity test will be in clinical use next year (Kimball Genetics). It will test for variations in CYP2C9 and VKORC1
- An UGT1A1 gene variant is associated with leukopenia if prescribed camptosar, a drug for colon cancer (Oncoscreen)
- A TPMT variant is associated with slow metabolism of 6-mercaptopurine, used in the treatment of childhood leukemia and inflammatory bowel diseases (Pharmaco-Gendia)
- Epigenomics is currently developing tests based on DNA methylation
- Tyrosine kinase inhibitor gleevec inhibits the ABL, ARG, SCF/KIT, and PDGFRA and PDGFRB kinases in CML. Mutations in ABL can arise as secondary mutations in previously sensitive leukemias (Pharmaco-Gendia)
Needless to say that I have excluded here specific HIV mutations that may induce resistance to particular drugs (as I learned last week on a bioinformatics meeting here by Thomas Lengauer). I have also excluded all kind of sex-specific marker (e.g. SRY testing) and the whole nutrigenomics stuff.
Who knows more, for example about lansoprazole effectiveness, UGT1A9 and mycophenolic acid, UGT1A1 and irinotecan, COMT genotype and amphetamine response, pharmacogenetics of COX-2 inhibitors, and GRP78 responsiveness to chemotherapy? Is there any commercial test available for these genes? It seems that somebody should start a wiki on that, yea, yea.
Addendum 31-12-09
Here is another gene list; only 6 tests have been approved by the FDA; Nature reports about Oncotype DX and Prostate Px as well as MammaPrint. See also an UK based paper
HLA-B*5701 was most commonly tested to identify those at risk of abacavir hypersensitivity among patients with HIV. A number of barriers to testing were identified, including lack of clinician knowledge and a lack of scientific evidence.
INDELligent
I am detailing in a forthcoming paper in “Allergy”, that the contradicting results found with ADAM33 (the first positionally cloned asthma gene) probably results from a rather poor design of all follow-up studies.
It does not make so much sense to repeat over and over the same few SNP marker; instead a full resquencing of the linkage region would be necessary. From the analysis of public LD maps it is even possible that neighboring genes may be responsible for the observed associations.
I have also doubts if the SNP-centric view is always leading to success. BTW there is a new database of over 400,000 non-reduandt indels of which 280,000 are validated by comparison with other human or chimpanzee genomes (see Mills et al., the indels are available in dbSNP under the “Devine_lab” handle).
Blog together
Just read the entry of Janet D. Stemwedel about the 2007 North Carolina Science Blogging Conference, a “free, open and public event for scientists, educators, students, journalists, bloggers and anyone interested in discussing science communication, education and literacy on the Web.” Do not miss her tag “Shameless self-promotion” ;-) Three editors from The Lancet will take up to 49 registrants. Who can give me a free ride from Munich (bonus miles welcome)?
The journey is the reward
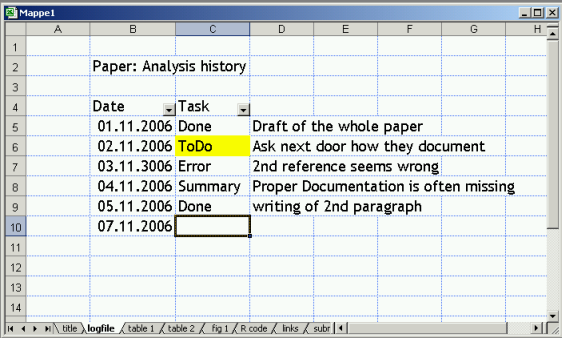
The analysis of a large dataset can be done in many different ways. At least in my experience documentation is getting confusing after many weeks of work – what has been done at which time? Can I reproduce my earlier findings? Where are the latest figures? What remains to be done? …
This is an ever increasing problem. Some information is documented in lab books, other in clinical journals, or even sitting on a server database where it may change over time. I have therefore described my own documentation procedure in a PDF paper that is online at this site.
With a quick view at the figure you will see that I am (mis-)using spreadsheets for documentation Please note (1) the drop down arrows at date and task that can be used for selecting lines even of long lists and (2) that tabs can used for fast switching between results (3) one tab contains all analysis scripts (4) and one tab all links.
Best of two worlds
Finally, linkage and association data can be used together after downloading new software using genotype inference.
It reduces the number of genotyping reactions and increases the power of genome-wide association studies. Our method combines sparse marker data from a linkage scan and high-resolution SNP genotypes for several individuals to infer genotypes for related individuals.
Sure, we
- could already test association only in linked families
- knew that linkage genome scans will improve the power of association
- could evaluate by stepc if a polymorphisms explains a linkage result
but this seems to be the best recycling for our old fashioned linkage data. Yea, yea.
Geo IP identification
The Geo IP database is available at Maxmind and allows to trace your home city from IP addresses. Here is a quick and dirty script to upload the Geo IP data into MySQL:
|wj_geo.cmd|
I would also put an index on loc_id. Finally the database should be available as
SELECT city
FROM GeoLiteCity INNER JOIN GeoLiteCityBlocks ON GeoLiteCity.locID = GeoLiteCityBlocks.locID
WHERE $myIP >= startIpNum AND $myIP <= endIpNum;
where $myIP is calculated as
substr($_SERVER['REMOTE_ADDR'],0,3) * 16777216 +
substr($_SERVER['REMOTE_ADDR'],4,3) * 65536 +
substr($_SERVER['REMOTE_ADDR'],8,3) * 256 +
substr($_SERVER['REMOTE_ADDR'],12,3)
A low-cost system for a PDF literature archiv I
Getting a scientific paper on your harddisk is quite simple. I am using a Fujitsu Scan Snap that can process a single page in a few seconds. The resulting PDF needs to be further tweaked by OCR recognition like ABBY FineReader (I couldn’t find any good open source alternative). FR will leave your PDF intact while adding recognized text as an overlay (or “underlay”). Unfortunately FR does not support batch processing but your OS will do by using a windows scripting engine like CLRscript. We also need a tool to extract a text file from the modified PDF. A good choice is pdftotext — look at the sourcecode and the DRM discussion before compiling it with a compiler like Cygwin. The following perl script doesn´t do anything than traversing your target directory and creating a batch file. As filenames offered by publishers are rather strange, I would first start to create some clean file names by replacing all spaces and brackets with something innocent like underscores.
perl.exe ocr.pl rename h:\pdf\2008\*.*
Now we create text files from the PDFs (usually done better by XPDF than directly by GDS).
perl.exe ocr.pl extract h:\pdf\2008\*.pdf
The resulting textfiles may be inspected: very small file sizes usually indicate no valid extraction and should be deleted before starting the OCR step as OCR is only done when text files are missing.
perl.exe ocr.pl ocr h:\pdf\2008\*.pdf
In the last step you may want to repeat the extract step.
ocr.zip
|wj_ocr.txt|
Unripe
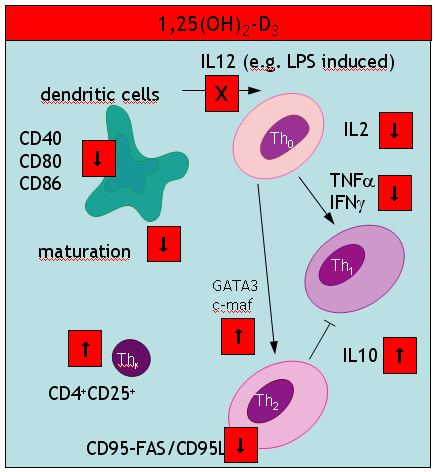
A key event in the vitamin D hypothesis of allergy induction is the immature state of dendritic cells. So far, maturity has been mainly described in terms of reduced expression of cell surface marker like CD80. A new study in Nature now further unravels how the capacity of DC to present antigen may be disturbed. Ubiquitination – the covalent attachment of ubiquitin polymers – of the MHC II ß chain ceases on maturation allowing the transport from endosomal compartments to the plasma membrane. Immature cells seem to be capable to some level of peptide-MHC interaction (at least for some selected antigens) although this process is greatly enhanced by maturation of DCs. Semi-maturity is believed to be an important inetrim stage where at least an earlier review argued
we propose a model in which steady-state migration and partial maturation (semi-maturation) of DCs is embedded as a major component within immune homeostasis, established for permanent and active tolerance induction against self-antigens derived from peripheral tissues by inducing antigen-specific CD4+ Tr cells. Semi-maturation induced by proinflammatory cytokines, such as TNF-alpha, seems to represent a unique developmental tolerogenic stage for DCs, which is based on the absence of proinflammatory cytokine production, despite high expression of MHC II and costimulatory molecules.
Another interesting study in the J Immunol – coined “alternatively activated dendritic cells” the authors probably talk about the same immature cells (compare with my cartoon summarizing a 2002 paper in Trend Mol Med). These immature DCs secrete high levels of IL10 (a paradox discussed in my most recent paper). In addition these DCs produce low amounts of IL12p70, TLR4 and CCR7. What was new to me, was an impressive list of pharmacological agents that suppress DC development: aspirin (also paracetamol?), corticosteroids, cyclosporine A, rapamycin (also other antibiotics?), and finally mycophenolate mofetil.
There is also an update of the IL10 paradox: Allergic sensitization may be down regulated by CD40 AGONISTs independent of IL10!
Finally, I would like to understand what immature really do after encountering allergen exposure. A new paper in nature immunology says that
immature DC are also thought to carry antigen to lymph nodes and to interact with naive T cells but without a previous maturation stimulus, those interactions result in abortive activation of the T cells, which can be eliminated, rendered unresponsive or induced to differentiate into regulatory T cells.
which still does not answer my question.
Vitamin D actions
Yea, yea.
Some are hybrids, some are not
Microchimerism is an interesting phenomenon that describes the hosting of foreign cells in an individuum – the prefix micro relates to the rather low counts of foreign cells (see the self discussion).
It is believed (but unproven) that most cases of microchimerism relate to the persistence of fetal cells in the maternal organism. The background of microchimerism is extremely complicated as highlighted in a recent review about the immunology of placentation in mammals. This paper has some nice cartoons about the types of placentation (epitheliochorial, endothelichorial and haemochorial) where the invasive potential of fetal trophoblast cells is the culprit of reciprocal (?) cell traffic between mother and fetus. The highest risk is found in women with induced abortion; cell count is ranging from 0 to 21 male cells per 100,000 female cells in peripheral blood; transfer may occur from mother <-> child, twin <-> twin, or sib <-> mother <-> sib.
Microchimerism has been examined in transplantation medicine (where the recipient replaces the outer donor organ epithelium), in blood transfusion and HCT, as well as in some autoimmune diseases (systemic sclerosis, SLE, thyroiditis, PBC). A clinical review reports that fetal cells have been found to persist for many years, probably for a lifetime.
I have doubts if that is true as I am not aware of any quantitative long-term study. Nearly all studies identified only male cells in women although now genomic studies of single cells are possible allowing a much better identification of foreign cells. If you are looking for a PhD thesis, microchimerism could be your field!
I already wondered if microchimerism could lead to genotyping errors, a question that can now easily be tested on the garbage of genotyping labs: We usually have genotyping errors in the 1-10 o/oo range; sometimes we see also triallelic SNPs. As far as I can renember, microchimerism has never been analyzed in the allergy field, although allergy can transplanted as well as asthma. Yea, yea.
Getting older – still the same?
Jonathan Keller is taking a picture every day since Oktober 1998. As seen on Youtube – with more series on the web. “Sie haben sich gar nicht verändert”. “Oh!”, sagte Herr Keuner und erbleichte (Bert Brecht).
Not just a face-lift
The redesign of the PLoS journals today is not just a facelift. They continue to have fresh ideas and unconventional approaches. A recent email said that
we encourage authors who are fluent in languages other than English to submit translated versions of an article summary … in other languages. Translations should be submitted as Supporting Information files labeled “Translation of the article summary (or entire article) into language XXX by author YYY”.
That is a nice idea, showing another benefit of online journals, yea, yea.
Are genes becoming more important with increasing age?
I found an apparent paradox between two studies. John Whitfield did a twin study in Australia and concluded that shared environmental effects decreased with age (from about 50% to 10%) while additive genetic effects increased. The new Sardinian study found higher heritabilities among younger individuals and explained that by an increase of environmental insults with age. Nice said, but who is right?